An integrated computational pipeline and database to support whole-genome sequence annotation
- PMID: 12537570
- PMCID: PMC151183
- DOI: 10.1186/gb-2002-3-12-research0081
An integrated computational pipeline and database to support whole-genome sequence annotation
Abstract
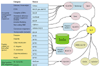
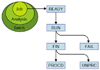
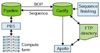
We describe here our experience in annotating the Drosophila melanogaster genome sequence, in the course of which we developed several new open-source software tools and a database schema to support large-scale genome annotation. We have developed these into an integrated and reusable software system for whole-genome annotation. The key contributions to overall annotation quality are the marshalling of high-quality sequences for alignments and the design of a system with an adaptable and expandable flexible architecture.
Figures
References
-
- Ensembl Analysis Pipeline http://www.ensembl.org/Docs/wiki/html/EnsemblDocs/Pipeline.html
-
- NCBI genome sequence and annotation process http://www.ncbi.nlm.nih.gov/genome/guide/build.html#annot
-
- Saccharomyces genome database http://genome-www.stanford.edu/Saccharomyces/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

