Annotation of the Drosophila melanogaster euchromatic genome: a systematic review
- PMID: 12537572
- PMCID: PMC151185
- DOI: 10.1186/gb-2002-3-12-research0083
Annotation of the Drosophila melanogaster euchromatic genome: a systematic review
Abstract
Background: The recent completion of the Drosophila melanogaster genomic sequence to high quality and the availability of a greatly expanded set of Drosophila cDNA sequences, aligning to 78% of the predicted euchromatic genes, afforded FlyBase the opportunity to significantly improve genomic annotations. We made the annotation process more rigorous by inspecting each gene visually, utilizing a comprehensive set of curation rules, requiring traceable evidence for each gene model, and comparing each predicted peptide to SWISS-PROT and TrEMBL sequences.
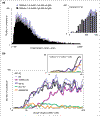
Results: Although the number of predicted protein-coding genes in Drosophila remains essentially unchanged, the revised annotation significantly improves gene models, resulting in structural changes to 85% of the transcripts and 45% of the predicted proteins. We annotated transposable elements and non-protein-coding RNAs as new features, and extended the annotation of untranslated (UTR) sequences and alternative transcripts to include more than 70% and 20% of genes, respectively. Finally, cDNA sequence provided evidence for dicistronic transcripts, neighboring genes with overlapping UTRs on the same DNA sequence strand, alternatively spliced genes that encode distinct, non-overlapping peptides, and numerous nested genes.
Conclusions: Identification of so many unusual gene models not only suggests that some mechanisms for gene regulation are more prevalent than previously believed, but also underscores the complex challenges of eukaryotic gene prediction. At present, experimental data and human curation remain essential to generate high-quality genome annotations.
Figures







References
-
- FlyBase: a database of the Drosophila Genome http://www.flybase.org
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Rubin GM, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, Harvey DA. A Drosophila complementary DNA resource. Science. 2000;287:2222–2224. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

