The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective
- PMID: 12537573
- PMCID: PMC151186
- DOI: 10.1186/gb-2002-3-12-research0084
The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective
Abstract
Background: Transposable elements are found in the genomes of nearly all eukaryotes. The recent completion of the Release 3 euchromatic genomic sequence of Drosophila melanogaster by the Berkeley Drosophila Genome Project has provided precise sequence for the repetitive elements in the Drosophila euchromatin. We have used this genomic sequence to describe the euchromatic transposable elements in the sequenced strain of this species.
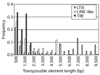
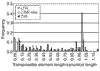
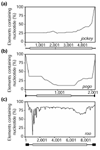
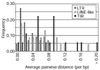
Results: We identified 85 known and eight novel families of transposable element varying in copy number from one to 146. A total of 1,572 full and partial transposable elements were identified, comprising 3.86% of the sequence. More than two-thirds of the transposable elements are partial. The density of transposable elements increases an average of 4.7 times in the centromere-proximal regions of each of the major chromosome arms. We found that transposable elements are preferentially found outside genes; only 436 of 1,572 transposable elements are contained within the 61.4 Mb of sequence that is annotated as being transcribed. A large proportion of transposable elements is found nested within other elements of the same or different classes. Lastly, an analysis of structural variation from different families reveals distinct patterns of deletion for elements belonging to different classes.
Conclusions: This analysis represents an initial characterization of the transposable elements in the Release 3 euchromatic genomic sequence of D. melanogaster for which comparison to the transposable elements of other organisms can begin to be made. These data have been made available on the Berkeley Drosophila Genome Project website for future analyses.
Figures


References
-
- Craig NL, Craigie R, Gellert M, Lambowitz AM, Eds Mobile DNA II Washington, DC: ASM Press; 2002.
-
- Green MM. Genetic instability in Drosophila melanogaster: Mutable miniature (mmu). Mutat Res. 1975;29:77–84. - PubMed
-
- Demerec M. Unstable genes. Bot Rev. 1935;1:233–248.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

