Systematic determination of patterns of gene expression during Drosophila embryogenesis
- PMID: 12537577
- PMCID: PMC151190
- DOI: 10.1186/gb-2002-3-12-research0088
Systematic determination of patterns of gene expression during Drosophila embryogenesis
Abstract
Background: Cell-fate specification and tissue differentiation during development are largely achieved by the regulation of gene transcription.
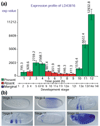
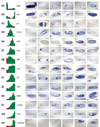
Results: As a first step to creating a comprehensive atlas of gene-expression patterns during Drosophila embryogenesis, we examined 2,179 genes by in situ hybridization to fixed Drosophila embryos. Of the genes assayed, 63.7% displayed dynamic expression patterns that were documented with 25,690 digital photomicrographs of individual embryos. The photomicrographs were annotated using controlled vocabularies for anatomical structures that are organized into a developmental hierarchy. We also generated a detailed time course of gene expression during embryogenesis using microarrays to provide an independent corroboration of the in situ hybridization results. All image, annotation and microarray data are stored in publicly available database. We found that the RNA transcripts of about 1% of genes show clear subcellular localization. Nearly all the annotated expression patterns are distinct. We present an approach for organizing the data by hierarchical clustering of annotation terms that allows us to group tissues that express similar sets of genes as well as genes displaying similar expression patterns.
Conclusions: Analyzing gene-expression patterns by in situ hybridization to whole-mount embryos provides an extremely rich dataset that can be used to identify genes involved in developmental processes that have been missed by traditional genetic analysis. Systematic analysis of rigorously annotated patterns of gene expression will complement and extend the types of analyses carried out using expression microarrays.
Figures






References
-
- Pollet N, Niehrs C. Expression profiling by systematic high-throughput in situ hybridization to whole-mount embryos. Methods Mol Biol. 2001;175:309–321. - PubMed
-
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21(Suppl):20–24. - PubMed
-
- Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. - PubMed
-
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21(Suppl):33–37. - PubMed
-
- Jones KW, Robertson FW. Localisation of reiterated nucleotide sequences in Drosophila and mouse by in situ hybridisation of complementary RNA. Chromosoma. 1970;31:331–345. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

