Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival
- PMID: 12540574
- PMCID: PMC145379
- DOI: 10.1128/IAI.71.2.922-929.2003
Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival
Abstract
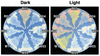
Pathogenic mycobacteria survive and replicate within host macrophages, but the molecular mechanisms involved in this necessary step in the pathogenesis of infection are not completely understood. Mycobacterium marinum has recently been used as a model for aspects of the pathogenesis of tuberculosis because of its close genetic relationship to Mycobacterium tuberculosis and because of similarities in the pathology and course of infection caused by this organism in its natural hosts, fish and frogs, with tuberculosis in humans. In order to advance the utility of the M. marinum model, we have developed efficient transposon mutagenesis of the organism by using a Drosophila melanogaster mariner-based transposon. To determine the efficiency of transposition, we have analyzed pigmentation mutants from the transposon mutant library. In addition to insertions in four known genes in the pathway of pigment biosynthesis, two insertions in novel genes were identified in our mutant library. One of these is in a putative inhibitor of the carotenoid biosynthesis pathway. The second unexpected insertion is in an intergenic region between two genes homologous to Rv2603c and Rv2604c of M. tuberculosis. In addition to a pigmentation defect, this mutant showed increased susceptibility to singlet oxygen and grew poorly in murine macrophages. Complementation with M. tuberculosis genomic DNA encompassing Rv2603c to Rv2606c corrected the pigmentation and growth defects of the mutant. These data demonstrate the utility of mariner-based transposon mutagenesis of M. marinum and that M. marinum can be used to study the function of M. tuberculosis genes involved in intracellular survival and replication.
Figures


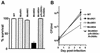

References
-
- Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. - PubMed
-
- Bessereau, J. L., A. Wright, D. C. Williams, K. Schuske, M. W. Davis, and E. M. Jorgensen. 2001. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413:70-74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

