Quantitation of changes in protein phosphorylation: a simple method based on stable isotope labeling and mass spectrometry
- PMID: 12540831
- PMCID: PMC298695
- DOI: 10.1073/pnas.232735599
Quantitation of changes in protein phosphorylation: a simple method based on stable isotope labeling and mass spectrometry
Abstract
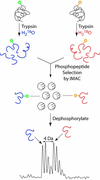
Reversible protein phosphorylation plays an important role in many cellular processes. However, a simple and reliable method to measure changes in the extent of phosphorylation is lacking. Here, we present a method to quantitate the changes in phosphorylation occurring in a protein in response to a stimulus. The method consists of three steps: (i) enzymatic digestion in H(2)16O or isotopically enriched H(2)18O to label individual pools of differentially phosphorylated proteins; (ii) affinity selection of phosphopeptides from the combined digests by immobilized metal-affinity chromatography; and (iii) dephosphorylation with alkaline phosphatase to allow for quantitation of changes of phosphorylation by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. We applied this strategy to the analysis of the yeast nitrogen permease reactivator protein kinase involved in the target of rapamycin signaling pathway. Alteration in the extent of phosphorylation at Ser-353 and Ser-357 could be easily assessed and quantitated both in wild-type yeast cells treated with rapamycin and in cells lacking the SIT4 phosphatase responsible for dephosphorylating nitrogen permease reactivator protein. The method described here is simple and allows quantitation of relative changes in the level of phosphorylation in signaling proteins, thus yielding information critical for understanding the regulation of complex protein phosphorylation cascades.
Figures




References
-
- McLachlin D T, Chait B T. Curr Opin Chem Biol. 2001;5:591–602. - PubMed
-
- Radimerski T, Mini T, Schneider U, Wettenhall R E, Thomas G, Jenoe P. Biochemistry. 2000;39:5766–5774. - PubMed
-
- Resing K A, Ahn N G. Methods Enzymol. 1997;283:29–44. - PubMed
-
- Goshe M B, Conrads T P, Panisko E A, Angell N H, Veenstra T D, Smith R D. Anal Chem. 2001;73:2578–2586. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

