An empirical correlation between secondary structure content and averaged chemical shifts in proteins
- PMID: 12547802
- PMCID: PMC1302698
- DOI: 10.1016/S0006-3495(03)74937-6
An empirical correlation between secondary structure content and averaged chemical shifts in proteins
Abstract
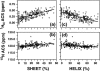
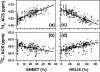
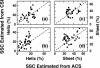
It is shown that the averaged chemical shift (ACS) of a particular nucleus in the protein backbone empirically correlates well to its secondary structure content (SSC). Chemical shift values of more than 200 proteins obtained from the Biological Magnetic Resonance Bank are used to calculate ACS values, and the SSC is estimated from the corresponding three-dimensional coordinates obtained from the Protein Data Bank. ACS values of (1)H(alpha) show the highest correlation to helical and sheet structure content (correlation coefficient of 0.80 and 0.75, respectively); (1)H(N) exhibits less reliability (0.65 for both sheet and helix), whereas such correlations are poor for the heteronuclei. SSC estimated using this correlation shows a good agreement with the conventional chemical shift index-based approach for a set of proteins that only have chemical shift information but no NMR or x-ray determined three-dimensional structure. These results suggest that even chemical shifts averaged over the entire protein retain significant information about the secondary structure. Thus, the correlation between ACS and SSC can be used to estimate secondary structure content and to monitor large-scale secondary structural changes in protein, as in folding studies.
Figures
Similar articles
-
Estimation of protein secondary structure content directly from NMR spectra using an improved empirical correlation with averaged chemical shift.J Struct Funct Genomics. 2005 Dec;6(4):281-5. doi: 10.1007/s10969-005-9002-8. Epub 2005 Nov 9. J Struct Funct Genomics. 2005. PMID: 16283427
-
Probability-based protein secondary structure identification using combined NMR chemical-shift data.Protein Sci. 2002 Apr;11(4):852-61. doi: 10.1110/ps.3180102. Protein Sci. 2002. PMID: 11910028 Free PMC article.
-
Toward direct determination of conformations of protein building units from multidimensional NMR experiments VI: chemical shift analysis of his to gain 3D structure and protonation state information.J Comput Chem. 2005 Oct;26(13):1307-17. doi: 10.1002/jcc.20266. J Comput Chem. 2005. PMID: 15999335
-
Use of chemical shifts and coupling constants in nuclear magnetic resonance structural studies on peptides and proteins.Methods Enzymol. 1994;239:392-416. doi: 10.1016/s0076-6879(94)39015-0. Methods Enzymol. 1994. PMID: 7830592 Review. No abstract available.
-
Protein chemical shift analysis: a practical guide.Biochem Cell Biol. 1998;76(2-3):153-63. doi: 10.1139/bcb-76-2-3-153. Biochem Cell Biol. 1998. PMID: 9923684 Review.
Cited by
-
Application of data mining tools for classification of protein structural class from residue based averaged NMR chemical shifts.Biochim Biophys Acta. 2015 Oct;1854(10 Pt A):1545-52. doi: 10.1016/j.bbapap.2015.02.016. Epub 2015 Mar 7. Biochim Biophys Acta. 2015. PMID: 25758094 Free PMC article.
-
The recognition of multi-class protein folds by adding average chemical shifts of secondary structure elements.Saudi J Biol Sci. 2016 Mar;23(2):189-97. doi: 10.1016/j.sjbs.2015.10.008. Epub 2015 Dec 11. Saudi J Biol Sci. 2016. PMID: 26980999 Free PMC article.
-
Characterization of protein secondary structure from NMR chemical shifts.Prog Nucl Magn Reson Spectrosc. 2009 Apr 5;54(3-4):141-165. doi: 10.1016/j.pnmrs.2008.06.002. Prog Nucl Magn Reson Spectrosc. 2009. PMID: 20160946 Free PMC article. No abstract available.
-
Estimation of protein secondary structure content directly from NMR spectra using an improved empirical correlation with averaged chemical shift.J Struct Funct Genomics. 2005 Dec;6(4):281-5. doi: 10.1007/s10969-005-9002-8. Epub 2005 Nov 9. J Struct Funct Genomics. 2005. PMID: 16283427
-
An evaluation of chemical shift index-based secondary structure determination in proteins: influence of random coil chemical shifts.J Biomol NMR. 2004 Oct;30(2):143-53. doi: 10.1023/b:jnmr.0000048940.51331.49. J Biomol NMR. 2004. PMID: 15666561
References
-
- Ando, I., S. Kuroki, H. Kurosu, and T. Yamanobe. 2001. NMR chemical shift calculations and structural characterizations of polymers. Progress in Nuclear Magnetic Resonance Spectroscopy. 39:79–133.
-
- Case, D. A., H. J. Dyson, and P. E. Wright. 1994. Use of chemical shifts and coupling constants in nuclear magnetic resonance structural studies of peptides and proteins. Methods Enzymol. 239:392–416. - PubMed
-
- Cornilescu, G., F. Delaglio, and A. Bax. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 13:289–302. - PubMed
-
- Dalgarno, D. C., B. A. Levine, and R. J. Williams. 1983. Structural information from NMR secondary chemical shifts of peptide alpha C-H protons in proteins. Biosci. Rep. 3:443–452. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

