Interaction of the equine herpesvirus 1 EICP0 protein with the immediate-early (IE) protein, TFIIB, and TBP may mediate the antagonism between the IE and EICP0 proteins
- PMID: 12552007
- PMCID: PMC141080
- DOI: 10.1128/jvi.77.4.2675-2685.2003
Interaction of the equine herpesvirus 1 EICP0 protein with the immediate-early (IE) protein, TFIIB, and TBP may mediate the antagonism between the IE and EICP0 proteins
Abstract
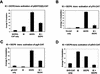
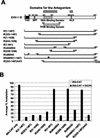
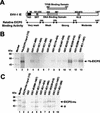
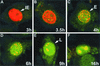
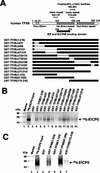
The equine herpesvirus 1 (EHV-1) immediate-early (IE) and EICP0 proteins are potent trans-activators of EHV-1 promoters; however, in transient-transfection assays, the IE protein inhibits the trans-activation function of the EICP0 protein. Assays with IE mutant proteins revealed that its DNA-binding domain, TFIIB-binding domain, and nuclear localization signal may be important for the antagonism between the IE and EICP0 proteins. In vitro interaction assays with the purified IE and EICP0 proteins indicated that these proteins interact directly. At late times postinfection, the IE and EICP0 proteins colocalized in the nuclei of infected equine cells. Transient-transfection assays showed that the EICP0 protein trans-activated EHV-1 promoters harboring only a minimal promoter region (TATA box and cap site), suggesting that the EICP0 protein trans-activates EHV-1 promoters by interactions with general transcription factor(s). In vitro interaction assays revealed that the EICP0 protein interacted directly with the basal transcription factors TFIIB and TBP and that the EICP0 protein (amino acids [aa] 143 to 278) mediated the interaction with aa 125 to 174 of TFIIB. Our unpublished data showed that the IE protein interacts with the same domain (aa 125 to 174) of TFIIB and with TBP. Taken together, these results suggested that interaction of the EICP0 protein with the IE protein, TFIIB, and TBP may mediate the antagonism between the IE and EICP0 proteins.
Figures

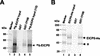

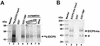

Similar articles
-
Full trans-activation mediated by the immediate-early protein of equine herpesvirus 1 requires a consensus TATA box, but not its cognate binding sequence.Virus Res. 2016 Jan 4;211:222-32. doi: 10.1016/j.virusres.2015.10.026. Epub 2015 Nov 2. Virus Res. 2016. PMID: 26541315 Free PMC article.
-
The unique IR2 protein of equine herpesvirus 1 negatively regulates viral gene expression.J Virol. 2006 May;80(10):5041-9. doi: 10.1128/JVI.80.10.5041-5049.2006. J Virol. 2006. PMID: 16641295 Free PMC article.
-
Characterization of the trans-activation properties of equine herpesvirus 1 EICP0 protein.J Virol. 2000 Feb;74(3):1200-8. doi: 10.1128/jvi.74.3.1200-1208.2000. J Virol. 2000. PMID: 10627530 Free PMC article.
-
A negative regulatory element (base pairs -204 to -177) of the EICP0 promoter of equine herpesvirus 1 abolishes the EICP0 protein's trans-activation of its own promoter.J Virol. 2004 Nov;78(21):11696-706. doi: 10.1128/JVI.78.21.11696-11706.2004. J Virol. 2004. PMID: 15479811 Free PMC article.
-
Transcriptional activation: enter TFIIB.Trends Biochem Sci. 1991 Sep;16(9):317-8. doi: 10.1016/0968-0004(91)90130-n. Trends Biochem Sci. 1991. PMID: 1949150 Review. No abstract available.
Cited by
-
Initial characterization of 17 viruses harboring mutant forms of the immediate-early gene of equine herpesvirus 1.Virus Genes. 2005 Oct;31(2):229-39. doi: 10.1007/s11262-005-1801-2. Virus Genes. 2005. PMID: 16025249
-
Full trans-activation mediated by the immediate-early protein of equine herpesvirus 1 requires a consensus TATA box, but not its cognate binding sequence.Virus Res. 2016 Jan 4;211:222-32. doi: 10.1016/j.virusres.2015.10.026. Epub 2015 Nov 2. Virus Res. 2016. PMID: 26541315 Free PMC article.
-
Equid herpesvirus type 1 activates platelets.PLoS One. 2015 Apr 23;10(4):e0122640. doi: 10.1371/journal.pone.0122640. eCollection 2015. PLoS One. 2015. PMID: 25905776 Free PMC article.
-
The equine herpesvirus-1 IR3 gene that lies antisense to the sole immediate-early (IE) gene is trans-activated by the IE protein, and is poorly expressed to a protein.Virology. 2007 Jun 20;363(1):15-25. doi: 10.1016/j.virol.2007.01.024. Epub 2007 Feb 15. Virology. 2007. PMID: 17306852 Free PMC article.
-
The unique IR2 protein of equine herpesvirus 1 negatively regulates viral gene expression.J Virol. 2006 May;80(10):5041-9. doi: 10.1128/JVI.80.10.5041-5049.2006. J Virol. 2006. PMID: 16641295 Free PMC article.
References
-
- Bruatowski, S. 1994. The basics of basal transcription by RNA polymerase II. Cell 77:1-3. - PubMed
-
- Caughman, G. B., J. Staczek, and D. J. O'Callaghan. 1985. Equine herpesvirus type 1 infected cell polypeptides: evidence for immediate-early/early/late regulation of viral gene expression. Virology 145:49-61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

