Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals
- PMID: 12552096
- PMCID: PMC149932
- DOI: 10.1073/pnas.0437640100
Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals
Abstract
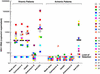
The presence of HIV-1 in latently infected, resting CD4(+) T cells has been clearly demonstrated in infected individuals; however, the extent of viral expression and the underlying mechanisms of the persistence of HIV-1 in this viral reservoir have not been fully delineated. Here, we show that resting CD4(+) T cells from the majority of viremic patients are capable of producing cell-free HIV-1 spontaneously ex vivo. The levels of HIV-1 released by resting CD4(+) T cells were not significantly reduced in the presence of inhibitors of cellular proliferation and viral replication. However, resting CD4(+) T cells from the majority of aviremic patients failed to produce virions, despite levels of HIV-1 proviral DNA and cell-associated HIV-1 RNA comparable to viremic patients. The DNA microarray analysis demonstrated that a number of genes involving transcription regulation, RNA processing and modification, and protein trafficking and vesicle transport were significantly upregulated in resting CD4(+) T cells of viremic patients compared to those of aviremic patients. These results suggest that active viral replication has a significant impact on the physiologic state of resting CD4(+) T cells in infected viremic patients and, in turn, allows release of HIV-1 without exogenous activation stimuli. In addition, given that no quantifiable virions were produced by the latent viral reservoir in the majority of aviremic patients despite the presence of cell-associated HIV-1 RNA, evidence for transcription of HIV-1 RNA in resting CD4(+) T cells of aviremic patients should not necessarily be taken as direct evidence for ongoing viral replication during effective therapy.
Figures



References
-
- Centers for Disease Control and Prevention. HIV/AIDS Surv Rep. 2000;12:1–44.
-
- Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, et al. Science. 1997;278:1295–1300. - PubMed
-
- Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Science. 1997;278:1291–1295. - PubMed
-
- Ho D D. Science. 1998;280:1866–1867. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

