Origin-specific unwinding of herpes simplex virus 1 DNA by the viral UL9 and ICP8 proteins: visualization of a specific preunwinding complex
- PMID: 12552114
- PMCID: PMC298698
- DOI: 10.1073/pnas.0237171100
Origin-specific unwinding of herpes simplex virus 1 DNA by the viral UL9 and ICP8 proteins: visualization of a specific preunwinding complex
Abstract
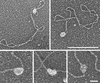
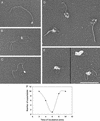
Herpes simplex virus 1 contains three origins of replication; two copies of oriS and one of a similar sequence, oriL. Here, the combined action of multiple factors known or thought to influence the opening of oriS are examined. These include the viral origin-binding protein, UL9, and single-strand binding protein ICP8, host cell topoisomerase I, and superhelicity of the DNA template. By using electron microscopy, it was observed that when ICP8 and UL9 proteins were added together to oriS-containing supertwisted DNA, a discrete preunwinding complex was formed at oriS on 40% of the molecules, which was shown by double immunolabeling electron microscopy to contain both proteins. This complex was relatively stable to extreme dilution. Addition of ATP led to the efficient unwinding of approximately 50% of the DNA templates. Unwinding proceeded until the acquisition of a high level of positive supertwists in the remaining duplex DNA inhibited further unwinding. Addition of topoisomerase I allowed further unwinding, opening >1 kb of DNA around oriS.
Figures


Similar articles
-
The herpes simplex virus type 1 origin-binding protein carries out origin specific DNA unwinding and forms stem-loop structures.EMBO J. 1996 Apr 1;15(7):1742-50. EMBO J. 1996. PMID: 8612599 Free PMC article.
-
An initial ATP-independent step in the unwinding of a herpes simplex virus type I origin of replication by a complex of the viral origin-binding protein and single-strand DNA-binding protein.Proc Natl Acad Sci U S A. 2001 Mar 13;98(6):3024-8. doi: 10.1073/pnas.061028298. Epub 2001 Mar 6. Proc Natl Acad Sci U S A. 2001. PMID: 11248025 Free PMC article.
-
[Complex of the herpes simplex virus initiator protein UL9 with DNA as a platform for the design of a new type of antiviral drugs].Biofizika. 2010 Mar-Apr;55(2):239-51. Biofizika. 2010. PMID: 20429277 Russian.
-
The herpes simplex virus type-1 single-strand DNA-binding protein, ICP8, increases the processivity of the UL9 protein DNA helicase.J Biol Chem. 1998 Jan 30;273(5):2676-83. doi: 10.1074/jbc.273.5.2676. J Biol Chem. 1998. PMID: 9446572
-
Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8.J Mol Biol. 1996 May 24;258(5):789-99. doi: 10.1006/jmbi.1996.0287. J Mol Biol. 1996. PMID: 8637010
Cited by
-
Evidence for DNA hairpin recognition by Zta at the Epstein-Barr virus origin of lytic replication.J Virol. 2010 Jul;84(14):7073-82. doi: 10.1128/JVI.02666-09. Epub 2010 May 5. J Virol. 2010. PMID: 20444899 Free PMC article.
-
Stepwise evolution of the herpes simplex virus origin binding protein and origin of replication.J Biol Chem. 2009 Jun 12;284(24):16246-16255. doi: 10.1074/jbc.M807551200. Epub 2009 Apr 7. J Biol Chem. 2009. PMID: 19351883 Free PMC article.
-
Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication.J Virol. 2004 Sep;78(17):9203-14. doi: 10.1128/JVI.78.17.9203-9214.2004. J Virol. 2004. PMID: 15308715 Free PMC article.
-
Association between the herpes simplex virus-1 DNA polymerase and uracil DNA glycosylase.J Biol Chem. 2010 Sep 3;285(36):27664-72. doi: 10.1074/jbc.M110.131235. Epub 2010 Jul 2. J Biol Chem. 2010. PMID: 20601642 Free PMC article.
-
Initiation of lytic DNA replication in Epstein-Barr virus: search for a common family mechanism.Future Virol. 2010 Jan;5(1):65-83. doi: 10.2217/fvl.09.69. Future Virol. 2010. PMID: 22468146 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

