Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype
- PMID: 12552116
- PMCID: PMC298726
- DOI: 10.1073/pnas.0335540100
Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype
Abstract
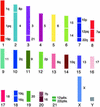
The Afrotheria, a supraordinal grouping of mammals whose radiation is rooted in Africa, is strongly supported by DNA sequence data but not by their disparate anatomical features. We have used flow-sorted human, aardvark, and African elephant chromosome painting probes and applied reciprocal painting schemes to representatives of two of the Afrotherian orders, the Tubulidentata (aardvark) and Proboscidea (elephants), in an attempt to shed additional light on the evolutionary affinities of this enigmatic group of mammals. Although we have not yet found any unique cytogenetic signatures that support the monophyly of the Afrotheria, embedded within the aardvark genome we find the strongest evidence yet of a mammalian ancestral karyotype comprising 2n = 44. This karyotype includes nine chromosomes that show complete conserved synteny to those of man, six that show conservation as single chromosome arms or blocks in the human karyotype but that occur on two different chromosomes in the ancestor, and seven neighbor-joining combinations (i.e., the synteny is maintained in the majority of species of the orders studied so far, but which corresponds to two chromosomes in humans). The comparative chromosome maps presented between human and these Afrotherian species provide further insight into mammalian genome organization and comparative genomic data for the Afrotheria, one of the four major evolutionary clades postulated for the Eutheria.
Figures




References
-
- Simpson G C. Bull Am Mus Nat Hist. 1945;85:1–350.
-
- Novacek M J. Nature. 1992;356:121–125. - PubMed
-
- Novacek M J. Curr Biol. 2001;11:573–575. - PubMed
-
- Springer M S, Cleven G C, Madsen O, de Yong W W, Waddell V G, Amrine H M, Stanhope M J. Nature. 1997;388:61–64. - PubMed
-
- Stanhope M J, Madsen O, Waddell V G, Cleven G C, de Jong W W, Springer M S. Mol Phylogenet Evol. 1998;9:501–508. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

