Genomic data sampling and its effect on classification performance assessment
- PMID: 12553886
- PMCID: PMC149349
- DOI: 10.1186/1471-2105-4-5
Genomic data sampling and its effect on classification performance assessment
Abstract
Background: Supervised classification is fundamental in bioinformatics. Machine learning models, such as neural networks, have been applied to discover genes and expression patterns. This process is achieved by implementing training and test phases. In the training phase, a set of cases and their respective labels are used to build a classifier. During testing, the classifier is used to predict new cases. One approach to assessing its predictive quality is to estimate its accuracy during the test phase. Key limitations appear when dealing with small-data samples. This paper investigates the effect of data sampling techniques on the assessment of neural network classifiers.
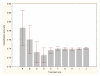
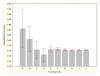
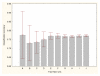
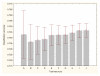
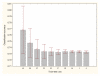
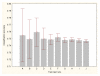
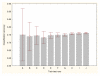
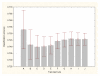
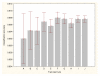
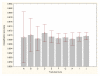
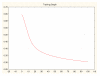
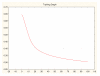
Results: Three data sampling techniques were studied: Cross-validation, leave-one-out, and bootstrap. These methods are designed to reduce the bias and variance of small-sample estimations. Two prediction problems based on small-sample sets were considered: Classification of microarray data originating from a leukemia study and from small, round blue-cell tumours. A third problem, the prediction of splice-junctions, was analysed to perform comparisons. Different accuracy estimations were produced for each problem. The variations are accentuated in the small-data samples. The quality of the estimates depends on the number of train-test experiments and the amount of data used for training the networks.
Conclusion: The predictive quality assessment of biomolecular data classifiers depends on the data size, sampling techniques and the number of train-test experiments. Conservative and optimistic accuracy estimations can be obtained by applying different methods. Guidelines are suggested to select a sampling technique according to the complexity of the prediction problem under consideration.
Figures




Similar articles
-
Ensemble Feature Learning of Genomic Data Using Support Vector Machine.PLoS One. 2016 Jun 15;11(6):e0157330. doi: 10.1371/journal.pone.0157330. eCollection 2016. PLoS One. 2016. PMID: 27304923 Free PMC article.
-
Accurate cancer classification using expressions of very few genes.IEEE/ACM Trans Comput Biol Bioinform. 2007 Jan-Mar;4(1):40-53. doi: 10.1109/TCBB.2007.1006. IEEE/ACM Trans Comput Biol Bioinform. 2007. PMID: 17277412
-
Mixture classification model based on clinical markers for breast cancer prognosis.Artif Intell Med. 2010 Feb-Mar;48(2-3):129-37. doi: 10.1016/j.artmed.2009.07.008. Epub 2009 Dec 14. Artif Intell Med. 2010. PMID: 20005686
-
Functional genomics and proteomics in the clinical neurosciences: data mining and bioinformatics.Prog Brain Res. 2006;158:83-108. doi: 10.1016/S0079-6123(06)58004-5. Prog Brain Res. 2006. PMID: 17027692 Review.
-
Support vector machine applications in bioinformatics.Appl Bioinformatics. 2003;2(2):67-77. Appl Bioinformatics. 2003. PMID: 15130823 Review.
Cited by
-
Assessment of Internal Validity of Prognostic Models through Bootstrapping and Multiple Imputation of Missing Data.Iran J Public Health. 2012;41(5):110-5. Epub 2012 May 31. Iran J Public Health. 2012. PMID: 23113185 Free PMC article.
-
Telediagnosis of Parkinson's disease using measurements of dysphonia.J Med Syst. 2010 Aug;34(4):591-9. doi: 10.1007/s10916-009-9272-y. Epub 2009 Mar 14. J Med Syst. 2010. PMID: 20703913
-
Challenges in the analysis of mass-throughput data: a technical commentary from the statistical machine learning perspective.Cancer Inform. 2007 Feb 16;2:133-62. Cancer Inform. 2007. PMID: 19458765 Free PMC article.
-
Multiclass classification of microarray data samples with a reduced number of genes.BMC Bioinformatics. 2011 Feb 22;12:59. doi: 10.1186/1471-2105-12-59. BMC Bioinformatics. 2011. PMID: 21342522 Free PMC article.
-
Predictive modeling using a somatic mutational profile in ovarian high grade serous carcinoma.PLoS One. 2013;8(1):e54089. doi: 10.1371/journal.pone.0054089. Epub 2013 Jan 10. PLoS One. 2013. PMID: 23326577 Free PMC article.
References
-
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Sampas N, Dougherty E, Wang E, Marincola F, Gooden C, Lueders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V, Hayward N, Trent J. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

