Molecular evidence for a positive role of Spt4 in transcription elongation
- PMID: 12554661
- PMCID: PMC140732
- DOI: 10.1093/emboj/cdg047
Molecular evidence for a positive role of Spt4 in transcription elongation
Abstract
We have previously shown that yeast mutants of the THO complex have a defect in gene expression, observed as an impairment of lacZ transcription. Here we analyze the ability of mutants of different transcription elongation factors to transcribe lacZ. We found that spt4Delta, like THO mutants, impaired transcription of lacZ and of long and GC-rich DNA sequences fused to the GAL1 promoter. Using a newly developed in vitro transcription elongation assay, we show that Spt4 is required in elongation. There is a functional interaction between Spt4 and THO, detected by the lethality or strong gene expression defect and hyper-recombination phenotypes of double mutants in the W303 genetic background. Our results indicate that Spt4-Spt5 has a positive role in transcription elongation and suggest that Spt4-Spt5 and THO act at different steps during mRNA biogenesis.
Figures
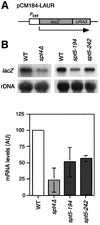
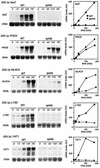
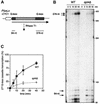



References
-
- Aso T., Lane,W.S., Conaway,J.W. and Conaway,R.C. (1995) Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science, 269, 1439–1443. - PubMed
-
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

