Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability
- PMID: 12554667
- PMCID: PMC140749
- DOI: 10.1093/emboj/cdg065
Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability
Abstract
Editing of misactivated amino acids by class I tRNA synthetases is encoded by a specialized internal domain specific to class I enzymes. In contrast, little is known about editing activities of the structurally distinct class II enzymes. Here we show that the class II alanyl-tRNA synthetase (AlaRS) has a specialized internal domain that appears weakly related to an appended domain of threonyl-tRNA synthetase (ThrRS), but is unrelated to that found in class I enzymes. Editing of misactivated glycine or serine was shown to require a tRNA cofactor. Specific mutations in the aforementioned domain disrupt editing and lead to production of mischarged tRNA. This class-specific editing domain was found to be essential for cell growth, in the presence of elevated concentrations of glycine or serine. In contrast to ThrRS, where the editing domain is not found in all three kingdoms of living organisms, it was incorporated early into AlaRSs and is present throughout evolution. Thus, tRNA-dependent editing by AlaRS may have been critical for making the genetic code sufficiently accurate to generate the tree of life.
Figures
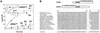
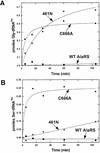


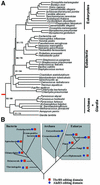

References
-
- Baldwin A.N. and Berg,P. (1966) Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J. Biol. Chem., 241, 839–845. - PubMed
-
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem., 72, 248–254. - PubMed
-
- Buechter D.D. and Schimmel,P. (1993) Dissection of a class II tRNA synthetase: determinants for minihelix recognition are tightly associated with domain for amino acid activation. Biochemistry, 32, 5267–5272. - PubMed
-
- Carter C.W. Jr (1993) Cognition, mechanism and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem., 62, 715–748. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

