The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth
- PMID: 12554669
- PMCID: PMC140753
- DOI: 10.1093/emboj/cdg069
The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth
Abstract
The translation initiation factor eIF4E is involved in the modulation of cellular growth. In the nucleus, where eIF4E is associated with PML nuclear bodies, eIF4E mediates nucleocytoplasmic transport of specific transcripts, and this contributes to its transformation activity. Surprisingly, we found that a trans cription factor, the proline-rich homeodomain protein PRH, is a negative regulator of eIF4E in myeloid cells, interacting with eIF4E through a conserved binding site typically found in translational regulators. Through this interaction, PRH inhibits eIF4E-dependent mRNA transport and subsequent transformation. These activities of PRH are independent of its transcriptional functions. Further, we found that 199 homeodomain proteins contain potential eIF4E-binding sites. Thus, there could be many tissue-specific regulators of eIF4E. These findings provide a model for regulation of a general factor, eIF4E, in tissue- specific contexts, and suggest that its regulation is important in differentiation and development.
Figures
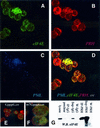


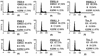
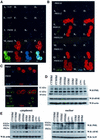
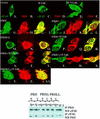


References
-
- Altmann M., Muller,P.P., Pelletier,J., Sonenberg,N. and Trachsel,H. (1989) A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J. Biol. Chem., 264, 12145–12147. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

