The alternating ATPase domains of MutS control DNA mismatch repair
- PMID: 12554674
- PMCID: PMC140748
- DOI: 10.1093/emboj/cdg064
The alternating ATPase domains of MutS control DNA mismatch repair
Abstract
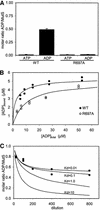
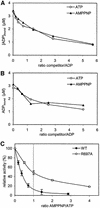
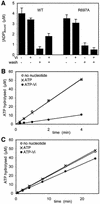
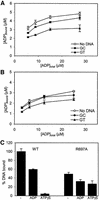
DNA mismatch repair is an essential safeguard of genomic integrity by removing base mispairings that may arise from DNA polymerase errors or from homologous recombination between DNA strands. In Escherichia coli, the MutS enzyme recognizes mismatches and initiates repair. MutS has an intrinsic ATPase activity crucial for its function, but which is poorly understood. We show here that within the MutS homodimer, the two chemically identical ATPase sites have different affinities for ADP, and the two sites alternate in ATP hydrolysis. A single residue, Arg697, located at the interface of the two ATPase domains, controls the asymmetry. When mutated, the asymmetry is lost and mismatch repair in vivo is impaired. We propose that asymmetry of the ATPase domains is an essential feature of mismatch repair that controls the timing of the different steps in the repair cascade.
Figures


References
-
- Abrahams J.P., Leslie,A.G., Lutter,R. and Walker,J.E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature, 370, 621–628. - PubMed
-
- Aquilina G. and Bignami,M. (2001) Mismatch repair in correction of replication errors and processing of DNA damage. J. Cell Physiol., 187, 145–154. - PubMed
-
- Bellacosa A. (2001) Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ., 8, 1076–1092. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

