Imaging the localized protein interactions between Pit-1 and the CCAAT/enhancer binding protein alpha in the living pituitary cell nucleus
- PMID: 12554785
- PMCID: PMC2910340
- DOI: 10.1210/me.2002-0136
Imaging the localized protein interactions between Pit-1 and the CCAAT/enhancer binding protein alpha in the living pituitary cell nucleus
Abstract
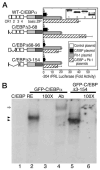
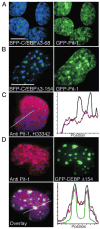
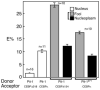
The homeodomain protein Pit-1 cooperates with the basic-leucine zipper protein CCAAT/enhancer binding protein alpha (C/EBPalpha) to control pituitary-specific prolactin gene transcription. We previously observed that C/EBPalpha was concentrated in regions of centromeric heterochromatin in pituitary GHFT1-5 cells and that coexpressed Pit-1 redistributed C/EBPalpha to the subnuclear sites occupied by Pit-1. Here, we used fluorescence resonance energy transfer microscopy to show that when C/EBPalpha was recruited by Pit-1, the average distance separating the fluorophores labeling the proteins was less than 7 nm. A mutation in the Pit-1 homeodomain, or truncation of the C/EBPalpha transactivation domain disrupted the redistribution of C/EBPalpha by Pit-1. Fluorescence resonance energy transfer analysis revealed that the mutant Pit-1 still associated with C/EBPalpha, and the truncated C/EBPalpha still associated with Pit-1, but these interactions were preferentially localized in regions of centromeric heterochromatin. In contrast, a truncation in C/EBPalpha that prevented DNA binding also blocked its association with Pit-1, suggesting that the binding of C/EBPalpha to DNA is a critical first step in specifying its association with Pit-1. These findings indicated that the protein domains that specify the interaction of Pit-1 and C/EBPalpha are separable from the protein domains that direct the positioning of the associated proteins within the nucleus. The intimate association of Pit-1 and C/EBPalpha at certain sites within the living cell nucleus could foster their combinatorial activities in the regulation of pituitary-specific gene expression.
Figures



Similar articles
-
A PIT-1 homeodomain mutant blocks the intranuclear recruitment of the CCAAT/enhancer binding protein alpha required for prolactin gene transcription.Mol Endocrinol. 2003 Feb;17(2):209-22. doi: 10.1210/me.2001-0222. Mol Endocrinol. 2003. PMID: 12554749 Free PMC article.
-
Dynamic interactions between Pit-1 and C/EBPalpha in the pituitary cell nucleus.Mol Cell Biol. 2006 Nov;26(21):8087-98. doi: 10.1128/MCB.02410-05. Epub 2006 Aug 14. Mol Cell Biol. 2006. PMID: 16908544 Free PMC article.
-
CCAAT/enhancer binding protein alpha uses distinct domains to prolong pituitary cells in the growth 1 and DNA synthesis phases of the cell cycle.BMC Cell Biol. 2002 Mar 21;3:6. doi: 10.1186/1471-2121-3-6. BMC Cell Biol. 2002. PMID: 11914124 Free PMC article.
-
Pit-1 and pituitary function.J Pediatr Endocrinol. 1993 Jul-Dec;6(3-4):229-33. doi: 10.1515/jpem.1993.6.3-4.229. J Pediatr Endocrinol. 1993. PMID: 7920987 Review.
-
POU domain factors in neural development.Adv Exp Med Biol. 1998;449:39-53. doi: 10.1007/978-1-4615-4871-3_4. Adv Exp Med Biol. 1998. PMID: 10026784 Review.
Cited by
-
Functional protein delivery into neurons using polymeric nanoparticles.J Biol Chem. 2009 Mar 13;284(11):6972-81. doi: 10.1074/jbc.M805956200. Epub 2009 Jan 7. J Biol Chem. 2009. PMID: 19129199 Free PMC article.
-
Nuclear localization and dynamic properties of the Marek's disease virus oncogene products Meq and Meq/vIL8.J Virol. 2006 Feb;80(3):1160-6. doi: 10.1128/JVI.80.3.1160-1166.2006. J Virol. 2006. PMID: 16414993 Free PMC article.
-
A PIT-1 homeodomain mutant blocks the intranuclear recruitment of the CCAAT/enhancer binding protein alpha required for prolactin gene transcription.Mol Endocrinol. 2003 Feb;17(2):209-22. doi: 10.1210/me.2001-0222. Mol Endocrinol. 2003. PMID: 12554749 Free PMC article.
-
ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation.Autoimmunity. 2008 May;41(4):253-71. doi: 10.1080/08916930802024202. Autoimmunity. 2008. PMID: 18432406 Free PMC article. Review.
-
Dynamic interactions between Pit-1 and C/EBPalpha in the pituitary cell nucleus.Mol Cell Biol. 2006 Nov;26(21):8087-98. doi: 10.1128/MCB.02410-05. Epub 2006 Aug 14. Mol Cell Biol. 2006. PMID: 16908544 Free PMC article.
References
-
- Gutierrez-Hartmann A. INSIGHT: Pit-1/GHF-1: a pituitary-specific transcription factor linking general signaling pathways to cell-specific gene expression. Mol Endocrinol. 1994;8:1447–1449. - PubMed
-
- Dasen JS, Rosenfeld MG. Combinatorial codes in signaling and synergy: lessons from pituitary development. Curr Opin Genet Dev. 1999;9:566–574. - PubMed
-
- Schaufele F. CCAAT/enhancer-binding protein α activation of the rat growth hormone promoter in pituitary progenitor GHFT1–5 cells. J Biol Chem. 1996;271:21484–21489. - PubMed
-
- Jacob KK, Stanley FM. CCAAT/enhancer-binding protein α is a physiological regulator of prolactin gene expression. Endocrinology. 1999;140:4542–4550. - PubMed
-
- Pombo A, Cuello P, Schul W, Yoon JB, Roeder RG, Cook PR, Murphy S. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17:1768–1778. - PMC - PubMed

