Structure and allosteric regulation of the alpha X beta 2 integrin I domain
- PMID: 12554829
- PMCID: PMC149926
- DOI: 10.1073/pnas.0237387100
Structure and allosteric regulation of the alpha X beta 2 integrin I domain
Abstract
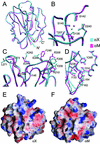
The integrin alpha X beta 2 (CD11c/CD18, p150,95) binds ligands through the I domain of the alpha X subunit. Ligands include the complement factor fragment iC3b, a key component in the innate immune defense, which, together with the expression of alpha X beta 2 on dendritic cells and on other leukocytes, suggests a role in the immune response. We now report the structure of the alpha X I domain resolved at 1.65 A by x-ray crystallography. To analyze structural requirements for ligand binding we made a mutation in the alpha X I domain C-terminal helix, which increased the affinity for iC3b approximately 200-fold to 2.4 microM compared with the wild-type domain affinity of approximately 400 microM. Gel permeation chromatography supported a conformational change between the wild-type and mutated domains. Conservation of allosteric regulation in the alpha X I domain points to the functional importance of this phenomenon.
Figures



Similar articles
-
An internal ligand-bound, metastable state of a leukocyte integrin, αXβ2.J Cell Biol. 2013 Nov 25;203(4):629-42. doi: 10.1083/jcb.201308083. J Cell Biol. 2013. PMID: 24385486 Free PMC article.
-
Multiple low-affinity interactions support binding of human osteopontin to integrin αXβ2.Biochim Biophys Acta. 2015 Aug;1854(8):930-8. doi: 10.1016/j.bbapap.2015.03.008. Epub 2015 Apr 1. Biochim Biophys Acta. 2015. PMID: 25839998
-
Characteristics of fibrinogen binding to the domain of CD11c, an alpha subunit of p150,95.Biochem Biophys Res Commun. 1999 Nov 2;264(3):630-4. doi: 10.1006/bbrc.1999.1564. Biochem Biophys Res Commun. 1999. PMID: 10543983
-
Conformational regulation of integrin structure and function.Annu Rev Biophys Biomol Struct. 2002;31:485-516. doi: 10.1146/annurev.biophys.31.101101.140922. Epub 2001 Oct 25. Annu Rev Biophys Biomol Struct. 2002. PMID: 11988479 Review.
-
CD11c (leukocyte integrin CR4 alpha subunit).J Biol Regul Homeost Agents. 1999 Apr-Jun;13(2):134-6. J Biol Regul Homeost Agents. 1999. PMID: 10503738 Review. No abstract available.
Cited by
-
Requirement of open headpiece conformation for activation of leukocyte integrin alphaXbeta2.Proc Natl Acad Sci U S A. 2010 Aug 17;107(33):14727-32. doi: 10.1073/pnas.1008663107. Epub 2010 Aug 2. Proc Natl Acad Sci U S A. 2010. PMID: 20679211 Free PMC article.
-
Two stage cadherin kinetics require multiple extracellular domains but not the cytoplasmic region.J Biol Chem. 2008 Jan 25;283(4):1848-56. doi: 10.1074/jbc.M708044200. Epub 2007 Nov 13. J Biol Chem. 2008. PMID: 17999960 Free PMC article.
-
Protein Binder (ProBi) as a New Class of Structurally Robust Non-Antibody Protein Scaffold for Directed Evolution.Viruses. 2021 Jan 27;13(2):190. doi: 10.3390/v13020190. Viruses. 2021. PMID: 33514045 Free PMC article.
-
The connection between metal ion affinity and ligand affinity in integrin I domains.Biochim Biophys Acta. 2007 Sep;1774(9):1148-55. doi: 10.1016/j.bbapap.2007.06.014. Epub 2007 Jul 12. Biochim Biophys Acta. 2007. PMID: 17702677 Free PMC article.
-
Identification of the target self-antigens in reperfusion injury.J Exp Med. 2006 Jan 23;203(1):141-52. doi: 10.1084/jem.20050390. Epub 2006 Jan 3. J Exp Med. 2006. PMID: 16390934 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

