Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding
- PMID: 12554879
- PMCID: PMC1370373
- DOI: 10.1261/rna.2131603
Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding
Abstract
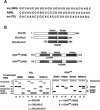
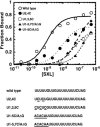
The molecular basis for specific recognition of simple homopolymeric sequences like the polypyrimidine tract (Py tract) by multiple RNA recognition motifs (RRMs) is not well understood. The Drosophila splicing repressor Sex lethal (SXL), which has two RRMs, can directly compete with the essential splicing factor U2AF(65), which has three RRMs, for binding to specific Py tracts. We have combined site-specific photocross-linking and chemical cleavage of the proteins to biochemically map cross-linking of each of the uracils within the Py tract to specific RRMs. For both proteins, RRM1 and RRM2 together constitute the minimal Py-tract recognition domain. The RRM3 of U2AF(65) shows no cross-linking to the Py tract. Both RRM1 and RRM2 of U2AF(65) and SXL can be cross-linked to certain residues, with RRM2 showing a surprisingly high number of residues cross-linked. The cross-linking data eliminate the possibility that shorter Py tracts are bound by fewer RRMs. We present a model to explain how the binding affinity can nonetheless change as a function of the length of the Py tract. The results indicate that multiple modes of binding result in an ensemble of RNA-protein complexes, which could allow tuning of the binding affinity without changing sequence specificity.
Figures






References
-
- Antson, A.A. 2000. Single-stranded-RNA binding proteins. Curr. Opin. Struct. Biol. 10: 87–94. - PubMed
-
- Bashaw, G.J. and Baker, B.S. 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789–798. - PubMed
-
- Black, D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367–370. - PubMed
-
- Burd, C.G. and Dreyfuss, G. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
