Sequence requirements for micro RNA processing and function in human cells
- PMID: 12554881
- PMCID: PMC1370375
- DOI: 10.1261/rna.2780503
Sequence requirements for micro RNA processing and function in human cells
Abstract
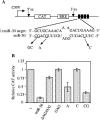
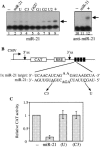
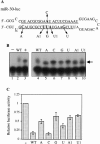
Most eukaryotes encode a substantial number of small noncoding RNAs termed micro RNAs (miRNAs). Previously, we have demonstrated that miR-30, a 22-nucleotide human miRNA, can be processed from a longer transcript bearing the proposed miR-30 stem-loop precursor and can translationally inhibit an mRNA-bearing artificial target sites. We also demonstrated that the miR-30 precursor stem can be substituted with a heterologous stem, which can be processed to yield novel miRNAs and can block the expression of endogenous mRNAs. Here, we show that a second human miRNA, termed miR-21, can also be effectively expressed when its precursor forms part of a longer mRNA. For both miR-30 and miR-21, mature miRNA production was highly dependent on the integrity of the precursor RNA stem, although the underlying sequence had little effect. In contrast, the sequence of the terminal loop affected miRNA production only moderately. Processing of the initial, miR-30-containing transcript led to the production of not only mature miR-30 but also to the largely nuclear excision of an approximately 65-nucleotide RNA that is likely to represent an important intermediate in miR-30 processing. Consistent with this hypothesis, mutations that affected mature miR-30 production inhibited expression of this miR-30 pre-miRNA to an equivalent degree. Although point mutations could block the ability of both miR-30 and miR-21 to inhibit the translation of mRNAs bearing multiple artificial miRNA target sites, single point mutations only attenuated the miRNA-mediated inhibition of genes bearing single, fully complementary targets. These results suggest that miRNAs, and the closely similar small interfering RNAs, cannot totally discriminate between RNA targets differing by a single nucleotide.
Figures




References
-
- Banerjee, D. and Slack, F. 2002. Control of developmental timing by small temporal RNAs: A paradigm for RNA-mediated regulation of gene expression. BioEssays 24: 119–129. - PubMed
-
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. - PubMed
-
- Bogerd, H.P., Fridell, R.A., Madore, S., and Cullen, B.R. 1995. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell 82: 485–494. - PubMed
-
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
