Effect of the pheromone-responsive G(alpha) and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase
- PMID: 12556475
- PMCID: PMC141143
- DOI: 10.1128/MCB.23.4.1135-1150.2003
Effect of the pheromone-responsive G(alpha) and phosphatase proteins of Saccharomyces cerevisiae on the subcellular localization of the Fus3 mitogen-activated protein kinase
Abstract
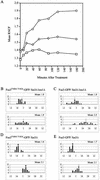
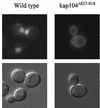
The mating-specific G(alpha) protein of Saccharomyces cerevisiae, Gpa1, stimulates adaptation to pheromone by a mechanism independent of G(beta gamma) sequestration. Genetic evidence suggests that Gpa1 targets the Fus3 mitogen-activated protein kinase, and it has recently been shown that the two proteins interact in cells responding to pheromone. To test the possibility that Gpa1 downregulates the mating signal by affecting the localization of Fus3, we created a Fus3-green fluorescent protein (GFP) fusion protein. In vegetative cells, Fus3-GFP was found in both the cytoplasm and the nucleus. Pheromone stimulated a measurable increase in the ratio of nuclear to cytoplasmic Fus3-GFP. In contrast, the relative level of nuclear Fus3-GFP decreased as cells recovered from pheromone arrest and did not increase when cells adapted to chronic stimulus were challenged again. Accumulation of Fus3-GFP in the nuclei of stimulated cells was also inhibited by overexpression of either wild-type Gpa1, the E364K hyperadaptive mutant form of Gpa1, or the Msg5 dually specific phosphatase. The effects of Gpa1 and Msg5 on Fus3 are partially interdependent. In a genetic screen for adaptive defective mutants, a nonsense allele of the nucleocytoplasmic transport receptor, Kap104, was identified. Truncation of the Kap104 cargo-binding domain blocked the effect of both Gpa1(E364K) and Msg5 on Fus3-GFP localization. Based on these results, we propose that Gpa1 and Msg5 work in concert to downregulate the mating signal and that they do so by inhibiting the pheromone-induced increase of Fus3 in the nucleus. Kap104 is required for the G(alpha)/phosphatase-mediated effect on Fus3 localization.
Figures





References
-
- Aitchison, J. D., G. Blobel, and M. P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624-627. - PubMed
-
- Alepuz, P. M., A. Jovanovic, V. Reiser, and G. Ammerer. 2001. Stress-induced MAP kinase Hog1 is part of transcription activation complexes. Mol. Cell 7:767-777. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
