Functional cross-antagonism between transcription factors FLI-1 and EKLF
- PMID: 12556498
- PMCID: PMC141137
- DOI: 10.1128/MCB.23.4.1390-1402.2003
Functional cross-antagonism between transcription factors FLI-1 and EKLF
Abstract
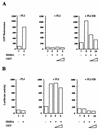
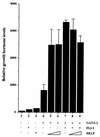
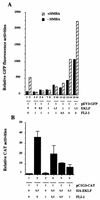
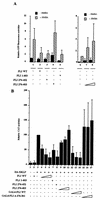
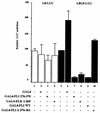
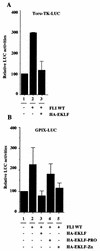
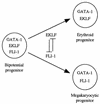
FLI-1 is an ETS family transcription factor which is overexpressed in Friend erythroleukemia and contributes to the blockage of differentiation of erythroleukemic cells. We show here that FLI-1 represses the transcriptional activity of the beta-globin gene promoter in MEL cells and interacts with two of its critical transactivators, GATA-1 and EKLF. Unexpectedly, FLI-1 enhances the stimulating activity of GATA-1 on a GATA-1-responsive promoter but represses that of EKLF on beta-globin and an EKLF-responsive artificial promoters. This repressive effect of FLI-1 requires the ETS DNA binding domain and its association with either the N- or C-terminal domain, which themselves interact with EKLF but not with GATA-1. Furthermore, the FLI-1 ETS domain alone behaves as an autonomous repression domain when linked to the Gal4 DNA binding domain. Taken together, these data indicate that FLI-1 represses EKLF-dependent transcription due to the repression activity of its ETS domain and its indirect recruitment to erythroid promoters by protein-protein interaction with EKLF. Reciprocally, we also show that EKLF itself represses the FLI-1-dependent megakaryocytic GPIX gene promoter, thus further suggesting that functional cross-antagonism between FLI-1 and EKLF might be involved in the control of the erythrocytic versus megakaryocytic differentiation of bipotential progenitors.
Figures

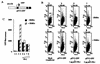



References
-
- Afrikanova, I., E. Yeh, D. Bartos, S. S. Watowich, and G. D. Longmore. 2002. Oncogene cooperativity in Friend erythroleukemia: erythropoietin receptor activation by the env gene of SFFV leads to transcriptional upregulation of PU.1, independent of SFFV proviral insertion. Oncogene 21:1272-1284. - PMC - PubMed
-
- Athanasiou, M., P. A. Clausen, G. J. Mavrothalassitis, X. K. Zhang, D. K. Watson, and D. G. Blair. 1996. Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell Growth Differ. 7:1525-1534. - PubMed
-
- Athanasiou, M., G. Mavrothalassitis, L. Sun-Hoffman, and D. G. Blair. 2000. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia 14:439-445. - PubMed
-
- Barnache, S., F. Wendling, C. Lacombe, N. Denis, M. Titeux, W. Vainchenker, and F. Moreau-Gachelin. 1998. Spi-1 transgenic mice develop a clonal erythroleukemia which does not depend on p53 mutation. Oncogene 16:2989-2995. - PubMed
-
- Barnache, S., P. Mayeux, B. Payrastre, and F. Moreau-Gachelin. 2001. Alterations of the phosphoinositide 3-kinase and mitogen-activated protein kinase signaling pathways in the erythropoietin-independent Spi-1/PU.1 transgenic proerythroblasts. Blood 98:2372-2381. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
