Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase
- PMID: 12556502
- PMCID: PMC141154
- DOI: 10.1128/MCB.23.4.1441-1452.2003
Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase
Abstract
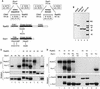
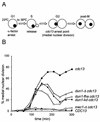
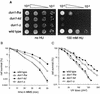
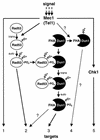
The serine-threonine kinase Dun1 contains a forkhead-associated (FHA) domain and functions in the DNA damage checkpoint pathway of Saccharomyces cerevisiae. It belongs to the Chk2 family of checkpoint kinases, which includes S. cerevisiae Rad53 and Mek1, Schizosaccharomyces pombe Cds1, and human Chk2. Dun1 is required for DNA damage-induced transcription of certain target genes, transient G(2)/M arrest after DNA damage, and DNA damage-induced phosphorylation of the DNA repair protein Rad55. Here we report that the FHA phosphoprotein recognition domain of Dun1 is required for direct phosphorylation of Dun1 by Rad53 kinase in vitro and in vivo. trans phosphorylation by Rad53 does not require the Dun1 kinase activity and is likely to involve only a transient interaction between the two kinases. The checkpoint functions of Dun1 kinase in DNA damage-induced transcription, G(2)/M cell cycle arrest, and Rad55 phosphorylation are severely compromised in an FHA domain mutant of Dun1. As a consequence, the Dun1 FHA domain mutant displays enhanced sensitivity to genotoxic stress induced by UV, methyl methanesulfonate, and the replication inhibitor hydroxyurea. We show that the Dun1 FHA domain is critical for direct kinase-to-kinase signaling from Rad53 to Dun1 in the DNA damage checkpoint pathway.
Figures





Similar articles
-
Phosphorylation of Sae2 Mediates Forkhead-associated (FHA) Domain-specific Interaction and Regulates Its DNA Repair Function.J Biol Chem. 2015 Apr 24;290(17):10751-63. doi: 10.1074/jbc.M114.625293. Epub 2015 Mar 11. J Biol Chem. 2015. PMID: 25762720 Free PMC article.
-
Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade.Mol Cell. 2008 Jun 20;30(6):767-78. doi: 10.1016/j.molcel.2008.05.013. Mol Cell. 2008. PMID: 18570878
-
Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae.J Biol Chem. 2007 Jan 12;282(2):986-95. doi: 10.1074/jbc.M609322200. Epub 2006 Nov 17. J Biol Chem. 2007. PMID: 17114794 Free PMC article.
-
Phosphatases, DNA damage checkpoints and checkpoint deactivation.Cell Cycle. 2007 Dec 15;6(24):3058-64. doi: 10.4161/cc.6.24.5100. Epub 2007 Sep 20. Cell Cycle. 2007. PMID: 18075314 Review.
-
Chk1 and Cds1: linchpins of the DNA damage and replication checkpoint pathways.J Cell Sci. 2000 Nov;113 ( Pt 22)(Pt 22):3889-96. doi: 10.1242/jcs.113.22.3889. J Cell Sci. 2000. PMID: 11058076 Free PMC article. Review.
Cited by
-
DNA damage checkpoints inhibit mitotic exit by two different mechanisms.Mol Cell Biol. 2007 Jul;27(14):5067-78. doi: 10.1128/MCB.00095-07. Epub 2007 May 7. Mol Cell Biol. 2007. PMID: 17485442 Free PMC article.
-
Mek1 stabilizes Hop1-Thr318 phosphorylation to promote interhomolog recombination and checkpoint responses during yeast meiosis.Nucleic Acids Res. 2012 Dec;40(22):11416-27. doi: 10.1093/nar/gks920. Epub 2012 Oct 9. Nucleic Acids Res. 2012. PMID: 23047948 Free PMC article.
-
Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways.Proc Natl Acad Sci U S A. 2003 May 27;100(11):6628-33. doi: 10.1073/pnas.1131932100. Epub 2003 May 5. Proc Natl Acad Sci U S A. 2003. PMID: 12732713 Free PMC article.
-
A proteome-wide analysis of kinase-substrate network in the DNA damage response.J Biol Chem. 2010 Apr 23;285(17):12803-12. doi: 10.1074/jbc.M110.106989. Epub 2010 Feb 27. J Biol Chem. 2010. PMID: 20190278 Free PMC article.
-
Yeast Dun1 Kinase Regulates Ribonucleotide Reductase Small Subunit Localization in Response to Iron Deficiency.J Biol Chem. 2016 Apr 29;291(18):9807-17. doi: 10.1074/jbc.M116.720862. Epub 2016 Mar 12. J Biol Chem. 2016. PMID: 26970775 Free PMC article.
References
-
- Ahn, J.-Y., X. Li, H. L. Davis, and C. E. Canman. 2002. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J. Biol. Chem. 277:19389-19395. - PubMed
-
- Alcasabas, A. A., A. J. Osborn, J. Bachant, F. H. Hu, P. J. H. Werler, K. Bousset, K. Furuya, J. F. X. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. - PubMed
-
- Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401-2415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
