Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae
- PMID: 12560485
- PMCID: PMC149208
- DOI: 10.1093/nar/gkg188
Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae
Abstract
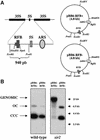
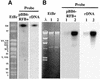
In the ribosomal DNA (rDNA) of Saccharomyces cerevisiae replication forks progressing against transcription stall at a polar replication fork barrier (RFB) located close to and downstream of the 35S transcription unit. Forks blocked at this barrier are potentially recombinogenic. Plasmids bearing the RFB sequence in its active orientation integrated into the chromosomal rDNA in sir2 mutant cells but not in wild-type cells, indicating that the histone deacetylase silencing protein Sir2 (Sir2p), which also modulates the aging process in yeast, suppresses the recombination competence of forks blocked at the rDNA RFB. Orientation of the RFB sequence in its inactive course or its abolition by FOB1 deletion avoided plasmid integration in sir2 mutant cells, indicating that stalling of the forks in the plasmid context was required for recombination to take place. Altogether these results strongly suggest that one of the functions of Sir2p is to modulate access of the recombination machinery to the forks stalled at the rDNA RFB.
Figures


References
-
- Bastia D. and Mohanty,B.K. (1996) Mechanisms for completing DNA replication. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, New York, NY, pp. 177–215.
-
- Hill T.M., Pelletier,A.J., Tecklenburg,M.L. and Kuempel,P.L. (1988) Identification of the DNA sequence from E. coli terminus region that halts replication forks. Cell, 55, 459–466. - PubMed
-
- Brewer B.J. (1988) When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell, 53, 679–686. - PubMed
-
- Olavarrieta L., Hernández,P., Krimer,D.B. and Schvartzman,J.B. (2002) DNA knotting caused by head-on collision of transcription and replication. J. Mol. Biol., 322, 1–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

