Molecular characterization of homozygous hereditary factor I deficiency
- PMID: 12562389
- PMCID: PMC1808620
- DOI: 10.1046/j.1365-2249.2003.02077.x
Molecular characterization of homozygous hereditary factor I deficiency
Abstract
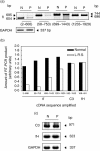
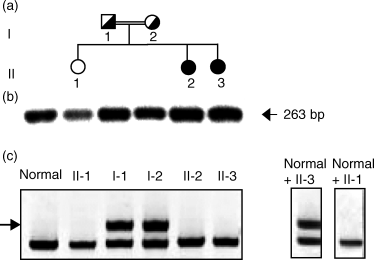
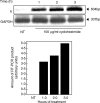
We have studied the molecular basis of factor I (fI) deficiency in two Brazilian sisters from a consanguineous family. By reverse transcription-polymerase chain reaction we observed that all fI cDNA amplified products from one sister had the same size as those of normal cDNA, however, they were significantly less intense. Sequencing analysis of subcloned cDNA revealed a dinucleotide insertion (AT) between positions 1204 and 1205 in the 11th exon that creates a stop codon 13 bp downstream of the insertion site. Genomic DNA sequencing and heteroduplex analysis confirmed that both probands are homozygous for this mutation, whereas their parents are heterozygous. The stop codon and the diminished amounts of fI cDNA could indicate increased fI mRNA instability, perhaps due to a mechanism of nonsense-mediated decay. This hypothesis is consistent with our observation that treatment with the translation inhibitor cycloheximide stabilized fI mRNA expression in proband's fibroblasts.
Figures

Similar articles
-
Homozygous hereditary C3 deficiency due to a premature stop codon.J Clin Immunol. 2002 Nov;22(6):321-30. doi: 10.1023/a:1020665614139. J Clin Immunol. 2002. PMID: 12462331
-
[Inherited afibrinogenemia caused by compound heterozygous mutations in the beta beta-chain of fibrinogen].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2005 Dec;13(6):1086-9. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2005. PMID: 16403286 Chinese.
-
Analysis of Iranian patients allowed the identification of the first truncating mutation in the fibrinogen Bbeta-chain gene causing afibrinogenemia.Haematologica. 2002 Aug;87(8):855-9. Haematologica. 2002. PMID: 12161363
-
A novel frameshift mutation in FGA accounting for congenital afibrinogenemia predicted to encode an aberrant peptide terminating 158 amino acids downstream.Blood Coagul Fibrinolysis. 2009 Jul;20(5):385-7. doi: 10.1097/MBC.0b013e328329f2a0. Blood Coagul Fibrinolysis. 2009. PMID: 19417632 Review.
-
The molecular basis of inherited afibrinogenaemia.Thromb Haemost. 2001 Jul;86(1):154-63. Thromb Haemost. 2001. PMID: 11487003 Review.
Cited by
-
Predicting physiological aging rates from a range of quantitative traits using machine learning.Aging (Albany NY). 2021 Oct 29;13(20):23471-23516. doi: 10.18632/aging.203660. Epub 2021 Oct 29. Aging (Albany NY). 2021. PMID: 34718232 Free PMC article.
-
Classical and Non-classical Presentations of Complement Factor I Deficiency: Two Contrasting Cases Diagnosed via Genetic and Genomic Methods.Front Immunol. 2019 Jun 7;10:1150. doi: 10.3389/fimmu.2019.01150. eCollection 2019. Front Immunol. 2019. PMID: 31231365 Free PMC article.
-
Complete Complement Factor I (CFI) deficiency: a systematic review of forty-nine patients including three novel cases.BMC Immunol. 2025 Jul 26;26(1):54. doi: 10.1186/s12865-025-00739-y. BMC Immunol. 2025. PMID: 40713518 Free PMC article.
-
Complement factor I deficiency: a not so rare immune defect: characterization of new mutations and the first large gene deletion.Orphanet J Rare Dis. 2012 Jun 18;7:42. doi: 10.1186/1750-1172-7-42. Orphanet J Rare Dis. 2012. PMID: 22710145 Free PMC article.
-
Rapid genome sequencing identifies novel variants in complement factor I.Cold Spring Harb Mol Case Stud. 2022 Dec 28;8(7):a006239. doi: 10.1101/mcs.a006239. Print 2022 Dec. Cold Spring Harb Mol Case Stud. 2022. PMID: 36577522 Free PMC article.
References
-
- Hebell T, Ahearn JM, Fearon DT. Suppression of the immune response by a soluble complement receptor of B lymphocytes. Science. 1991;254:102–5. - PubMed
-
- Dempsey PW, Allison MED, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science. 1996;271:348–50. - PubMed
-
- Vyse TJ, Späth PJ, Davies KA, Morley BJ, Philippe P, Athanassiou P, Giles CM, Walport MJ. Hereditary complement factor I deficiency. Q J Med. 1994;87:385–402. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

