Interactions between phage-shock proteins in Escherichia coli
- PMID: 12562786
- PMCID: PMC142853
- DOI: 10.1128/JB.185.4.1174-1180.2003
Interactions between phage-shock proteins in Escherichia coli
Abstract
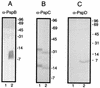
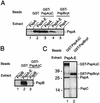
Expression of the pspABCDE operon of Escherichia coli is induced upon infection by filamentous phage and by many other stress conditions, including defects in protein export. Expression of the operon requires the alternative sigma factor sigma54 and the transcriptional activator PspF. In addition, PspA plays a negative regulatory role, and the integral-membrane proteins PspB and PspC play a positive one. In this study, we investigated whether the suggested protein-protein interactions implicated in this complex regulatory network can indeed be demonstrated. Antisera were raised against PspB, PspC, and PspD, which revealed, in Western blotting experiments, that PspC forms stable sodium dodecyl sulfate-resistant dimers and that the hypothetical pspD gene is indeed expressed in vivo. Fractionation experiments showed that PspD localizes as a peripherally bound inner membrane protein. Cross-linking studies with intact cells revealed specific interactions of PspA with PspB and PspC, but not with PspD. Furthermore, affinity-chromatography suggested that PspB could bind PspA only in the presence of PspC. These data indicate that regulation of the psp operon is mediated via protein-protein interactions.
Figures




References
-
- Adams, H., W. Teertstra, M. Koster, and J. Tommassen. 2002. PspE (phage-shock protein E) of Escherichia coli is a rhodanese. FEBS Lett. 518:173-176. - PubMed
-
- Agterberg, M., H. Adriaanse, A. van Bruggen, M. Karperien, and J. Tommassen. 1990. Outer-membrane PhoE protein of Escherichia coli K-12 as an exposure vector: possibilities and limitations. Gene 88:37-45. - PubMed
-
- Akrim, M., M. Bally, G. Ball, J. Tommassen, H. Teerink, A. Filloux, and A. Lazdunski. 1993. Xcp-mediated protein secretion in Pseudomonas aeruginosa: identification of two additional genes and evidence for regulation of xcp gene expression. Mol. Microbiol. 10:431-443. - PubMed
-
- Ansorge, W. 1985. Fast and sensitive detection of protein and DNA bands by treatment with potassium permanganate. J. Biochem. Biophys. Methods 11:13-20. - PubMed
-
- Bauer, M. F., C. Sirrenberg, W. Neupert, and M. Brunner. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87:33-41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

