Characterization of extradiol dioxygenases from a polychlorinated biphenyl-degrading strain that possess higher specificities for chlorinated metabolites
- PMID: 12562795
- PMCID: PMC142886
- DOI: 10.1128/JB.185.4.1253-1260.2003
Characterization of extradiol dioxygenases from a polychlorinated biphenyl-degrading strain that possess higher specificities for chlorinated metabolites
Abstract
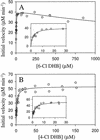
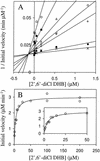
Recent studies demonstrated that 2,3-dihydroxybiphenyl 1,2-dioxygenase from Burkholderia sp. strain LB400 (DHBDLB400; EC 1.13.11.39) cleaves chlorinated 2,3-dihydroxybiphenyls (DHBs) less specifically than unchlorinated DHB and is competitively inhibited by 2',6'-dichloro-2,3-dihydroxybiphenyl (2',6'-diCl DHB). To determine whether these are general characteristics of DHBDs, we characterized DHBDP6-I and DHBDP6-III, two evolutionarily divergent isozymes from Rhodococcus globerulus strain P6, another good polychlorinated biphenyl (PCB) degrader. In contrast to DHBDLB400, both rhodococcal enzymes had higher specificities for some chlorinated DHBs in air-saturated buffer. Thus, DHBDP6-I cleaved the DHBs in the following order of specificity: 6-Cl DHB > 3'-Cl DHB approximately DHB approximately 4'-Cl DHB > 2'-Cl DHB > 4-Cl DHB > 5-Cl DHB. It also cleaved its preferred substrate, 6-Cl DHB, three times more specifically than DHB. Interestingly, some of the worst substrates for DHBDP6-I were among the best for DHBDP6-III (4-Cl DHB > 5-Cl DHB approximately 6-Cl DHB approximately 3'-Cl DHB > DHB > 2'-Cl DHB approximately 4'-Cl DHB; DHBDP6-III cleaved 4-Cl DHB two times more specifically than DHB). Generally, each of the monochlorinated DHBs inactivated the enzymes more rapidly than DHB. The exceptions were 4-Cl DHB for DHBDP6-I and 2'-Cl DHB for DHBDP6-III. As observed in DHBDLB400, chloro substituents influenced the reactivity of the dioxygenases with O2. For example, the apparent specificities of DHBDP6-I and DHBDP6-III for O2 in the presence of 2'-Cl DHB were lower than those in the presence of DHB by factors of >60 and 4, respectively. DHBDP6-I and DHBDP6-III shared the relative inability of DHBDLB400 to cleave 2',6'-diCl DHB (apparent catalytic constants of 0.088 +/- 0.004 and 0.069 +/- 0.002 s(-1), respectively). However, these isozymes had remarkably different apparent K(m) values for this compound (0.007 +/- 0.001, 0.14 +/- 0.01, and 3.9 +/- 0.4 micro M for DHBDLB400, DHBDP6-I, and DHBDP6-III, respectively). The markedly different reactivities of DHBDP6-I and DHBDP6-III with chlorinated DHBs undoubtedly contribute to the PCB-degrading activity of R. globerulus P6.
Figures
References
-
- Abramowicz, D. A. 1990. Aerobic and anaerobic biodegradation of PCBs: a review. Crit. Rev. Biotechnol. 10:241-251.
-
- Arciero, D. M., and J. D. Lipscomb. 1986. Binding of 17O-labeled substrate and inhibitors to protocatechuate 4,5-dioxygenase-nitrosyl complex. Evidence for direct substrate binding to the active site Fe2+ of extradiol dioxygenases. J. Biol. Chem. 261:2170-2178. - PubMed
-
- Arciero, D. M., A. M. Orville, and J. D. Lipscomb. 1985. [17O]Water and nitric oxide binding by protocatechuate 4,5-dioxygenase and catechol 2,3-dioxygenase. Evidence for binding of exogenous ligands to the active site Fe2+ of extradiol dioxygenases. J. Biol. Chem. 260:14035-14044. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

