Molecular characteristics of spontaneous deletions in the hyperthermophilic archaeon Sulfolobus acidocaldarius
- PMID: 12562797
- PMCID: PMC142876
- DOI: 10.1128/JB.185.4.1266-1272.2003
Molecular characteristics of spontaneous deletions in the hyperthermophilic archaeon Sulfolobus acidocaldarius
Abstract
Prokaryotic genomes acquire and eliminate blocks of DNA sequence by lateral gene transfer and spontaneous deletion, respectively. The basic parameters of spontaneous deletion, which are expected to influence the course of genome evolution, have not been determined for any hyperthermophilic archaeon. We therefore screened a number of independent pyrimidine auxotrophs of Sulfolobus acidocaldarius for deletions and sequenced those detected. Deletions accounted for only 0.4% of spontaneous pyrE mutations, corresponding to a frequency of about 10(-8) per cell. Nucleotide sequence analysis of five independent deletions showed no significant association of the endpoints with short direct repeats, despite the fact that several such repeats occur within the pyrE gene and that duplication mutations in pyrE reverted at high frequencies. Endpoints of the spontaneous deletions did not coincide with short inverted repeats or potential stem-loop structures. No consensus sequence common to all the deletions could be identified, although two deletions showed the potential of being stabilized by octanucleotide sequences elsewhere in pyrE, and another pair of deletions shared an octanucleotide at their 3' ends. The unusually low frequency and low sequence dependence of spontaneous deletions in the S. acidocaldarius pyrE gene compared to other genetic systems could not be explained in terms of possible constraints imposed by the 5-fluoroorotate selection.
Figures

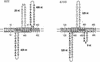
References
-
- Andersson, J. O., and S. G. E. Andersson. 1999. Genome degradation is an ongoing process in Rickettsia. Mol. Biol. Evol. 16:1178-1191. - PubMed
-
- Buhler, C., J. H. G. Lebbink, C. Bocs, R. Ladenstein, and P. Forterre. 2001. DNA topoisomerase VI generates ATP-dependent double-strand breaks with two-nucleotide overhangs. J. Biol. Chem. 276:37215-37222. - PubMed
-
- Bullock, P., J. J. Champoux, and M. Botchan. 1985. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science 230:954-958. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

