Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus
- PMID: 12562798
- PMCID: PMC142869
- DOI: 10.1128/JB.185.4.1273-1283.2003
Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus
Abstract
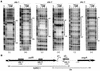
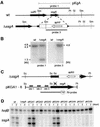
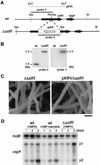
A-factor (2-isocapryloyl-3R-hydroxymethyl-gamma-butyrolactone) triggers morphological development and secondary metabolism in Streptomyces griseus. A transcriptional activator (AdpA) in the A-factor regulatory cascade switches on a number of genes required for both processes. AdBS11 was identified in a library of the DNA fragments that are bound by AdpA and mapped upstream of ssgA, which is essential for septum formation in aerial hyphae. Gel mobility shift assays and DNase I footprinting revealed three AdpA-binding sites at nucleotide positions about -235 (site 1), -110 (site 2), and +60 (site 3) with respect to the transcriptional start point, p1, of ssgA. ssgA had two transcriptional start points, one starting at 124 nucleotides (p1) and the other starting at 79 nucleotides (p2) upstream of the start codon of ssgA. Of the three binding sites, only sites 1 and 2 were required for transcriptional activation of p1 and p2 by AdpA. The transcriptional switch on of ssgA required the extracytoplasmic function sigma factor, sigma(AdsA), in addition to AdpA. However, it was unlikely that sigma(AdsA) recognized the two ssgA promoters, since their -35 and -10 sequences were not similar to the promoter sequence motifs recognized by sigma(BldN), a sigma(AdsA) homologue of Streptomyces coelicolor A3(2). An ssgA disruptant formed aerial hyphae, but did not form spores, irrespective of the carbon source of the medium, which indicated that ssgA is a member of the whi genes. Transcriptional analysis of ssfR, located just upstream of ssgA and encoding an IclR-type transcriptional regulator, suggested that no read-through from ssfR into ssgA occurred, and ssgA was transcribed in the absence of ssfR. ssgA was thus found to be controlled by AdpA and not by SsfR to a detectable extent. SsfR appeared to regulate spore septum formation independently of SsgA or through interaction with SsgA in some unknown way, because an ssfR disruptant also showed a whi phenotype.
Figures





Similar articles
-
An A-factor-dependent extracytoplasmic function sigma factor (sigma(AdsA)) that is essential for morphological development in Streptomyces griseus.J Bacteriol. 2000 Aug;182(16):4596-605. doi: 10.1128/JB.182.16.4596-4605.2000. J Bacteriol. 2000. PMID: 10913094 Free PMC article.
-
amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus.Mol Microbiol. 2003 Nov;50(4):1173-87. doi: 10.1046/j.1365-2958.2003.03760.x. Mol Microbiol. 2003. PMID: 14622407
-
Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus.J Bacteriol. 2002 Nov;184(21):6016-25. doi: 10.1128/JB.184.21.6016-6025.2002. J Bacteriol. 2002. PMID: 12374836 Free PMC article.
-
AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus.Biosci Biotechnol Biochem. 2005 Mar;69(3):431-9. doi: 10.1271/bbb.69.431. Biosci Biotechnol Biochem. 2005. PMID: 15784968 Review.
-
A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus.Front Biosci. 2002 Oct 1;7:d2045-57. doi: 10.2741/A897. Front Biosci. 2002. PMID: 12165483 Review.
Cited by
-
Complex structure of the DNA-binding domain of AdpA, the global transcription factor in Streptomyces griseus, and a target duplex DNA reveals the structural basis of its tolerant DNA sequence specificity.J Biol Chem. 2013 Oct 25;288(43):31019-29. doi: 10.1074/jbc.M113.473611. Epub 2013 Sep 9. J Biol Chem. 2013. PMID: 24019524 Free PMC article.
-
Transcriptional analysis of the cell division-related ssg genes in Streptomyces coelicolor reveals direct control of ssgR by AtrA.Antonie Van Leeuwenhoek. 2015 Jul;108(1):201-13. doi: 10.1007/s10482-015-0479-2. Epub 2015 May 23. Antonie Van Leeuwenhoek. 2015. PMID: 26002075 Free PMC article.
-
Control of the Streptomyces Subtilisin inhibitor gene by AdpA in the A-factor regulatory cascade in Streptomyces griseus.J Bacteriol. 2006 Sep;188(17):6207-16. doi: 10.1128/JB.00662-06. J Bacteriol. 2006. PMID: 16923887 Free PMC article.
-
The SsgA-like proteins in actinomycetes: small proteins up to a big task.Antonie Van Leeuwenhoek. 2008 Jun;94(1):85-97. doi: 10.1007/s10482-008-9225-3. Epub 2008 Feb 14. Antonie Van Leeuwenhoek. 2008. PMID: 18273689 Free PMC article. Review.
-
The Agrobacterium tumefaciens transcription factor BlcR is regulated via oligomerization.J Biol Chem. 2011 Jun 10;286(23):20431-40. doi: 10.1074/jbc.M110.196154. Epub 2011 Apr 4. J Biol Chem. 2011. PMID: 21467043 Free PMC article.
References
-
- Ando, N., K. Ueda, and S. Horinouchi. 1997. A Streptomyces griseus gene (sgaA) suppresses the growth disturbance caused by high osmolality and a high concentration of A-factor during early growth. Microbiology 143:2715-2723. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327-336. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

