Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa
- PMID: 12562802
- PMCID: PMC142842
- DOI: 10.1128/JB.185.4.1316-1325.2003
Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa
Abstract
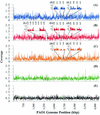
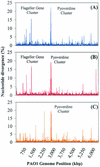
Whole-genome shotgun sequencing was used to study the sequence variation of three Pseudomonas aeruginosa isolates, two from clonal infections of cystic fibrosis patients and one from an aquatic environment, relative to the genomic sequence of reference strain PAO1. The majority of the PAO1 genome is represented in these strains; however, at least three prominent islands of PAO1-specific sequence are apparent. Conversely, approximately 10% of the sequencing reads derived from each isolate fail to align with the PAO1 backbone. While average sequence variation among all strains is roughly 0.5%, regions of pronounced differences were evident in whole-genome scans of nucleotide diversity. We analyzed two such divergent loci, the pyoverdine and O-antigen biosynthesis regions, by complete resequencing. A thorough analysis of isolates collected over time from one of the cystic fibrosis patients revealed independent mutations resulting in the loss of O-antigen synthesis alternating with a mucoid phenotype. Overall, we conclude that most of the PAO1 genome represents a core P. aeruginosa backbone sequence while the strains addressed in this study possess additional genetic material that accounts for at least 10% of their genomes. Approximately half of these additional sequences are novel.
Figures



References
-
- Belanger, M., L. L. Burrows, and J. S. Lam. 1999. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology 145:3505-3521. - PubMed
-
- Burns, J. L., R. L. Gibson, S. McNamara, D. Yim, J. Emerson, M. Rosenfeld, P. Hiatt, K. McCoy, R. Castile, A. L. Smith, and B. W. Ramsey. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444-452. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

