ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942
- PMID: 12562813
- PMCID: PMC142860
- DOI: 10.1128/JB.185.4.1415-1422.2003
ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942
Abstract
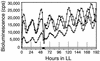
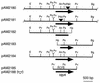
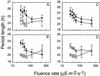
We generated random transposon insertion mutants to identify genes involved in light input pathways to the circadian clock of the cyanobacterium Synechococcus elongatus PCC 7942. Two mutants, AMC408-M1 and AMC408-M2, were isolated that responded to a 5-h dark pulse differently from the wild-type strain. The two mutants carried independent transposon insertions in an open reading frame here named ldpA (for light-dependent period). Although the mutants were isolated by a phase shift screening protocol, the actual defect is a conditional alteration in the circadian period. The mutants retain the wild-type ability to phase shift the circadian gene expression (bioluminescent reporter) rhythm if the timing of administration of the dark pulse is corrected for a 1-h shortening of the circadian period in the mutant. Further analysis indicated that the conditional short-period mutant phenotype results from insensitivity to light gradients that normally modulate the circadian period in S. elongatus, lengthening the period at low light intensities. The ldpA gene encodes a polypeptide that predicts a 7Fe-8S cluster-binding motif expected to be involved in redox reactions. We suggest that the LdpA protein modulates the circadian clock as an indirect function of light intensity by sensing changes in cellular physiology.
Figures
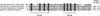


References
-
- Andersson, C. R., N. F. Tsinoremas, J. Shelton, N. V. Lebedeva, J. Yarrow, H. Min, S. S. Golden, H. Iwasaki, A. Tanabe, C. H. Johnson, and T. Kondo. 2000. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 305:527-542. - PubMed
-
- Aono, S., S. Nakamura, R. Aono, and I. Okura. 1994. Cloning and expression of the gene encoding the 7Fe type ferredoxin from a thermophilic hydrogen oxidizing bacterium, Bacillus schlegelii. Biochem. Biophys. Res. Commun. 201:938-942. - PubMed
-
- Aschoff, J. 1981. Freerunning and entrained circadian rhythms, p. 547-548. In J. Aschoff (ed.), Handbook of behavioral neurobiology, vol. 4. Biological rhythms. Plenum Press, New York, N.Y.
-
- Beinert, H., R. H. Holm, and E. Munck. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653-659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

