Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis
- PMID: 12562814
- PMCID: PMC142859
- DOI: 10.1128/JB.185.4.1423-1431.2003
Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis
Abstract
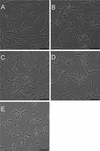
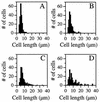
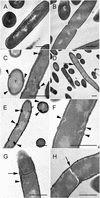
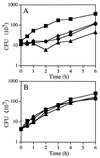
Penicillin-binding proteins (PBPs) catalyze the final, essential reactions of peptidoglycan synthesis. Three classes of PBPs catalyze either trans-, endo-, or carboxypeptidase activities on the peptidoglycan peptide side chains. Only the class A high-molecular-weight PBPs have clearly demonstrated glycosyltransferase activities that polymerize the glycan strands, and in some species these proteins have been shown to be essential. The Bacillus subtilis genome sequence contains four genes encoding class A PBPs and no other genes with similarity to their glycosyltransferase domain. A strain lacking all four class A PBPs has been constructed and produces a peptidoglycan wall with only small structural differences from that of the wild type. The growth rate of the quadruple mutant is much lower than those of strains lacking only three of the class A PBPs, and increases in cell length and frequencies of wall abnormalities were noticeable. The viability and wall production of the quadruple-mutant strain indicate that a novel enzyme can perform the glycosyltransferase activity required for peptidoglycan synthesis. This activity was demonstrated in vitro and shown to be sensitive to the glycosyltransferase inhibitor moenomycin. In contrast, the quadruple-mutant strain was resistant to moenomycin in vivo. Exposure of the wild-type strain to moenomycin resulted in production of a phenotype similar to that of the quadruple mutant.
Figures

References
-
- Adam, M., C. Fraipont, N. Rhazi, M. Nguyen-Disteche, B. Lakaye, J. M. Frere, B. Devreese, J. Van Beeumen, Y. van Heijenoort, J. van Heijenoort, and J. M. Ghuysen. 1997. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia coli catalyzes peptide bond formation from thiolesters and does not catalyze glycan chain polymerization from the lipid II intermediate. J. Bacteriol. 179:6005-6009. - PMC - PubMed
-
- Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, D.C.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

