Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus
- PMID: 12562815
- PMCID: PMC142846
- DOI: 10.1128/JB.185.4.1432-1442.2003
Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus
Abstract
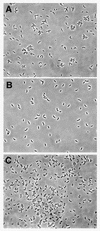
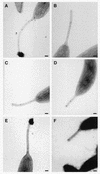
Adhesion to both abiotic and biotic surfaces by the gram-negative prothescate bacterium Caulobacter crescentus is mediated by a polar organelle called the "holdfast," which enables the bacterium to form stable monolayer biofilms. The holdfast, a complex polysaccharide composed in part of N-acetylglucosamine, localizes to the tip of the stalk (a thin cylindrical extension of the cell wall and membranes). We report here the isolation of adhesion mutants with transposon insertions in an uncharacterized gene cluster involved in holdfast biogenesis (hfs) as well as in previously identified polar development genes (podJ and pleC), and the holdfast attachment genes (hfa). Clean deletions of three of the four genes in the hfs gene cluster (hfsDAB) resulted in a severe holdfast biogenesis phenotype. These mutants do not bind to surfaces or to a fluorescently labeled lectin, specific for N-acetylglucosamine. Transmission electron microscopy indicated that the hfsDAB mutants fail to synthesize a holdfast at the stalk tip. The predicted hfs gene products have significant sequence similarity to proteins necessary for exopolysaccharide export in gram-negative bacteria. HfsA has sequence similarity to GumC from Xanthomonas campestris, which is involved in exopolysaccharide export in the periplasm. HfsD has sequence similarity to Wza from Escherichia coli, an outer membrane protein involved in secretion of polysaccharide through the outer membrane. HfsB is a novel protein involved in holdfast biogenesis. These data suggest that the hfs genes play an important role in holdfast export.
Figures



Similar articles
-
Mutations in Sugar-Nucleotide Synthesis Genes Restore Holdfast Polysaccharide Anchoring to Caulobacter crescentus Holdfast Anchor Mutants.J Bacteriol. 2018 Jan 10;200(3):e00597-17. doi: 10.1128/JB.00597-17. Print 2018 Feb 1. J Bacteriol. 2018. PMID: 29158242 Free PMC article.
-
The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk.Mol Microbiol. 2003 Sep;49(6):1671-83. doi: 10.1046/j.1365-2958.2003.03664.x. Mol Microbiol. 2003. PMID: 12950929
-
A Multiprotein Complex Anchors Adhesive Holdfast at the Outer Membrane of Caulobacter crescentus.J Bacteriol. 2019 Aug 22;201(18):e00112-19. doi: 10.1128/JB.00112-19. Print 2019 Sep 15. J Bacteriol. 2019. PMID: 31061167 Free PMC article.
-
Scaling down for a broader understanding of underwater adhesives - a case for the Caulobacter crescentus holdfast.Soft Matter. 2016 Nov 16;12(45):9132-9141. doi: 10.1039/c6sm02163h. Soft Matter. 2016. PMID: 27812588 Review.
-
Advantages and mechanisms of polarity and cell shape determination in Caulobacter crescentus.Curr Opin Microbiol. 2007 Dec;10(6):630-7. doi: 10.1016/j.mib.2007.09.007. Epub 2007 Nov 9. Curr Opin Microbiol. 2007. PMID: 17997127 Free PMC article. Review.
Cited by
-
Mutations in Sugar-Nucleotide Synthesis Genes Restore Holdfast Polysaccharide Anchoring to Caulobacter crescentus Holdfast Anchor Mutants.J Bacteriol. 2018 Jan 10;200(3):e00597-17. doi: 10.1128/JB.00597-17. Print 2018 Feb 1. J Bacteriol. 2018. PMID: 29158242 Free PMC article.
-
Development of surface adhesion in Caulobacter crescentus.J Bacteriol. 2004 Mar;186(5):1438-47. doi: 10.1128/JB.186.5.1438-1447.2004. J Bacteriol. 2004. PMID: 14973013 Free PMC article.
-
Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus.J Bacteriol. 2006 Oct;188(19):6841-50. doi: 10.1128/JB.00111-06. J Bacteriol. 2006. PMID: 16980487 Free PMC article.
-
Adhesins Involved in Attachment to Abiotic Surfaces by Gram-Negative Bacteria.Microbiol Spectr. 2015 Aug;3(4):10.1128/microbiolspec.MB-0018-2015. doi: 10.1128/microbiolspec.MB-0018-2015. Microbiol Spectr. 2015. PMID: 26350310 Free PMC article. Review.
-
Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development.J Bacteriol. 2006 Jul;188(14):5315-8. doi: 10.1128/JB.01725-05. J Bacteriol. 2006. PMID: 16816207 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1989. Current protocols in molecular biology. John Wiley/Greene, New York, N.Y.
-
- Brun, Y., G. Marczynski, and L. Shapiro. 1994. The expression of asymmetry during cell differentiation. Annu. Rev. Biochem. 63:419-450. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

