Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis
- PMID: 12566400
- PMCID: PMC420377
- DOI: 10.1101/gr.905003
Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis
Abstract
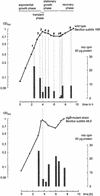
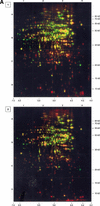
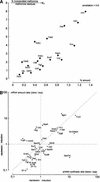
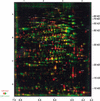
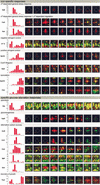
Dual channel imaging and warping of two-dimensional (2D) protein gels were used to visualize global changes of the gene expression patterns in growing Bacillus subtilis cells during entry into the stationary phase as triggered by glucose exhaustion. The 2D gels only depict single moments during the cells' growth cycle, but a sequential series of overlays obtained at specific points of the growth curve facilitates visualization of the developmental processes at the proteomics scale. During glucose starvation a substantial reprogramming of the protein synthesis pattern was found, with 150 proteins synthesized de novo and cessation of the synthesis of almost 400 proteins. Proteins induced following glucose starvation belong to two main regulation groups: general stress/starvation responses induced by different stresses or starvation stimuli (sigma(B)-dependent general stress regulon, stringent response, sporulation), and glucose-starvation-specific responses (drop in glycolysis, utilization of alternative carbon sources, gluconeogenesis). Using the dual channel approach, it was not only possible to identify those regulons or stimulons, but also to follow the fate of each single protein by the three-color code: red, newly induced but not yet accumulated; yellow, synthesized and accumulated; and green, still present, but no longer being synthesized. These green proteins, which represent a substantial part of the protein pool in the nongrowing cell, are not accessible by using DNA arrays. The combination of 2D gel electrophoresis and MALDI TOF mass spectrometry with the dual channel imaging technique provides a new and comprehensive view of the physiology of growing or starving bacterial cell populations, here for the case of the glucose-starvation response.
Figures





References
-
- Antelmann H., Bernhardt, J., Schmid, R., Mach, H., Völker, U., and Hecker, M. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18: 1451-1463. - PubMed
-
- Antelmann H., Tjalsma, H., Voigt, B., Ohlmeier, S., Bron, S., van Dijl, J.M., and Hecker, M. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11: 1484-1502. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
