Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network
- PMID: 12566402
- PMCID: PMC420374
- DOI: 10.1101/gr.234503
Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network
Abstract
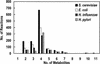
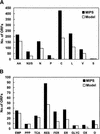
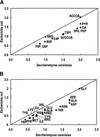
The metabolic network in the yeast Saccharomyces cerevisiae was reconstructed using currently available genomic, biochemical, and physiological information. The metabolic reactions were compartmentalized between the cytosol and the mitochondria, and transport steps between the compartments and the environment were included. A total of 708 structural open reading frames (ORFs) were accounted for in the reconstructed network, corresponding to 1035 metabolic reactions. Further, 140 reactions were included on the basis of biochemical evidence resulting in a genome-scale reconstructed metabolic network containing 1175 metabolic reactions and 584 metabolites. The number of gene functions included in the reconstructed network corresponds to approximately 16% of all characterized ORFs in S. cerevisiae. Using the reconstructed network, the metabolic capabilities of S. cerevisiae were calculated and compared with Escherichia coli. The reconstructed metabolic network is the first comprehensive network for a eukaryotic organism, and it may be used as the basis for in silico analysis of phenotypic functions.
Figures

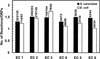
References
-
- André B. 1995. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 11: 1575-1611. - PubMed
-
- Bertsimas D. and Tsitsiklis, J.N., 1997. Linear optimization. Athena Scientific, Belmont, MA.
-
- Costanzo M.C., Crawford, M.E., Hirschman, J.E., Kranz, J.E., Olsen, P., Robertson, L.S., Skrzypek, M.S., Braun, B.R., Hopkins, K.L., Kondu, P., et al. 2001. YPD, PombePD and WormPD: Model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 29: 75-79. - PMC - PubMed
-
- Covert M.W., Schilling, C.H., Famili, I., Edwards, J.S., Goryanin, I.I., Selkov, E., and Palsson, B.Ø. 2001. Metabolic modeling of microbial strains in silico. Trends Biochem. Sci. 26: 179-186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
