The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast
- PMID: 12566428
- PMCID: PMC2172672
- DOI: 10.1083/jcb.200209022
The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast
Abstract
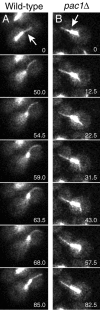
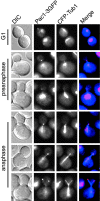
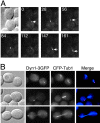
During mitosis in Saccharomyces cerevisiae, the mitotic spindle moves into the mother-bud neck via dynein-dependent sliding of cytoplasmic microtubules along the cortex of the bud. Here we show that Pac1, the yeast homologue of the human lissencephaly protein LIS1, plays a key role in this process. First, genetic interactions placed Pac1 in the dynein/dynactin pathway. Second, cells lacking Pac1 failed to display microtubule sliding in the bud, resulting in defective mitotic spindle movement and nuclear segregation. Third, Pac1 localized to the plus ends (distal tips) of cytoplasmic microtubules in the bud. This localization did not depend on the dynein heavy chain Dyn1. Moreover, the Pac1 fluorescence intensity at the microtubule end was enhanced in cells lacking dynactin or the cortical attachment molecule Num1. Fourth, dynein heavy chain Dyn1 also localized to the tips of cytoplasmic microtubules in wild-type cells. Dynein localization required Pac1 and, like Pac1, was enhanced in cells lacking the dynactin component Arp1 or the cortical attachment molecule Num1. Our results suggest that Pac1 targets dynein to microtubule tips, which is necessary for sliding of microtubules along the bud cortex. Dynein must remain inactive until microtubule ends interact with the bud cortex, at which time dynein and Pac1 appear to be offloaded from the microtubule to the cortex.
Figures





Comment in
-
LIS1 at the microtubule plus end and its role in dynein-mediated nuclear migration.J Cell Biol. 2003 Feb 3;160(3):289-90. doi: 10.1083/jcb.200212168. J Cell Biol. 2003. PMID: 12566423 Free PMC article. Review.
References
-
- Beach, D.L., J. Thibodeaux, P. Maddox, E. Yeh, and K. Bloom. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10:1497–1506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

