Complete genome sequence of Lactobacillus plantarum WCFS1
- PMID: 12566566
- PMCID: PMC149946
- DOI: 10.1073/pnas.0337704100
Complete genome sequence of Lactobacillus plantarum WCFS1
Abstract
The 3,308,274-bp sequence of the chromosome of Lactobacillus plantarum strain WCFS1, a single colony isolate of strain NCIMB8826 that was originally isolated from human saliva, has been determined, and contains 3,052 predicted protein-encoding genes. Putative biological functions could be assigned to 2,120 (70%) of the predicted proteins. Consistent with the classification of L. plantarum as a facultative heterofermentative lactic acid bacterium, the genome encodes all enzymes required for the glycolysis and phosphoketolase pathways, all of which appear to belong to the class of potentially highly expressed genes in this organism, as was evident from the codon-adaptation index of individual genes. Moreover, L. plantarum encodes a large pyruvate-dissipating potential, leading to various end-products of fermentation. L. plantarum is a species that is encountered in many different environmental niches, and this flexible and adaptive behavior is reflected by the relatively large number of regulatory and transport functions, including 25 complete PTS sugar transport systems. Moreover, the chromosome encodes >200 extracellular proteins, many of which are predicted to be bound to the cell envelope. A large proportion of the genes encoding sugar transport and utilization, as well as genes encoding extracellular functions, appear to be clustered in a 600-kb region near the origin of replication. Many of these genes display deviation of nucleotide composition, consistent with a foreign origin. These findings suggest that these genes, which provide an important part of the interaction of L. plantarum with its environment, form a lifestyle adaptation region in the chromosome.
Figures
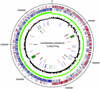
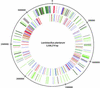
References
-
- Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Lancet. 2001;357:1076–1079. - PubMed
-
- Stiles M E, Holzapfel W H. Int J Food Microbiol. 1997;36:1–29. - PubMed
-
- Ahrne S, Nobaek S, Jeppsson B, Adlerberth I, Wold A E, Molin G. J Appl Microbiol. 1998;85:88–94. - PubMed
-
- Adawi D, Ahrne S, Molin G. Int J Food Microbiol. 2001;70:213–220. - PubMed
-
- Cunningham-Rundles S, Ahrne S, Bengmark S, Johann-Liang R, Marshall F, Metakis L, Califano C, Dunn A M, Grassey C, Hinds G, Cervia J. Am J Gastroenterol. 2000;95:22–25. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

