HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family
- PMID: 12566578
- PMCID: PMC141207
- DOI: 10.1105/tpc.006999
HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family
Abstract
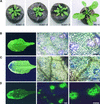
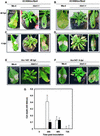
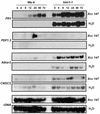
The hypersensitive response (HR) in plants is a programmed cell death that is commonly associated with disease resistance. A novel mutation in Arabidopsis, hlm1, which causes aberrant regulation of cell death, manifested by a lesion-mimic phenotype and an altered HR, segregated as a single recessive allele. Broad-spectrum defense mechanisms remained functional or were constitutive in the mutant plants, which also exhibited increased resistance to a virulent strain of Pseudomonas syringae pv tomato. In response to avirulent strains of the same pathogen, the hlm1 mutant showed differential abilities to restrict bacterial growth, depending on the avirulence gene expressed by the pathogen. The HLM1 gene encodes a cyclic nucleotide-gated channel, CNGC4. Preliminary study of the HLM1/CNGC4 gene pro-duct in Xenopus oocytes (inside-out patch-clamp technique) showed that CNGC4 is permeable to both K(+) and Na(+) and is activated by both cGMP and cAMP. HLM1 gene expression is induced in response to pathogen infection and some pathogen-related signals. Thus, HLM1 might constitute a common downstream component of the signaling pathways leading to HR/resistance.
Figures





References
-
- Anderson, K.V. (2000). Toll signaling pathways in the innate immune response. Curr. Opin. Immunol. 12, 13–19. - PubMed
-
- Baillieul, F., Genetet, I., Kopp, M., Saindrenan, P., Fritig, B., and Kauffman, S. (1995). A new elicitor of the hypersensitive response in tobacco: A fungal glycoprotein elicits cell death, expression of defense genes, production of salicylic acid, and induction of systemic acquired resistance. Plant J. 8, 551–560. - PubMed
-
- Baker, C.J., and Mock, N.M. (1994). An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell Tissue Organ Cult. 39, 7–12.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

