Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions
- PMID: 12566582
- PMCID: PMC141211
- DOI: 10.1105/tpc.006759
Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions
Abstract
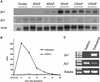
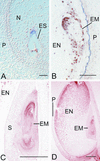
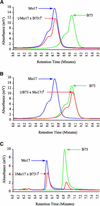
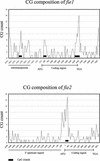
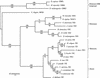
Two maize genes with predicted translational similarity to the Arabidopsis FIE (Fertilization-Independent Endosperm) protein, a repressor of endosperm development in the absence of fertilization, were cloned and analyzed. Genomic sequences of fie1 and fie2 show significant homology within coding regions but none within introns or 5' upstream. The fie1 gene is expressed exclusively in the endosperm of developing kernels starting at approximately 6 days after pollination. fie1 is an imprinted gene showing no detectable expression of the paternally derived fie1 allele during kernel development. Conversely, fie2 is expressed in the embryo sac before pollination. After pollination, its expression persists, predominantly in the embryo and at lower levels in the endosperm. The paternal fie2 allele is not expressed early in kernel development, but its transcription is activated at 5 days after pollination. fie2 is likely to be a functional ortholog of the Arabidopsis FIE gene, whereas fie1 has evolved a distinct function. The maize FIE2 and sorghum FIE proteins form a monophyletic group, sharing a closer relationship to each other than to the FIE1 protein, suggesting that maize fie genes originated from two different ancestral genomes.
Figures


References
-
- Alleman, M., and Doctor, J. (2000). Genomic imprinting in plants: Observation and evolutionary implications. Plant Mol. Biol. 43, 147–161. - PubMed
-
- Bradley, D., Carpenter, R., Somer, H., Hartley, N., and Coen, E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72, 85–95. - PubMed
-
- Brenner, S., et al. (2000. a). Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18, 630–634. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

