The biosynthesis of L-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis
- PMID: 12566589
- PMCID: PMC141218
- DOI: 10.1105/tpc.008425
The biosynthesis of L-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis
Abstract
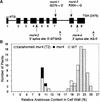
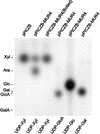
The mur4 mutant of Arabidopsis shows a 50% reduction in the monosaccharide L-Ara in leaf-derived cell wall material because of a partial defect in the 4-epimerization of UDP-D-Xyl to UDP-L-Ara. To determine the genetic lesion underlying the mur4 phenotype, the MUR4 gene was cloned by a map-based procedure and found to encode a type-II membrane protein with sequence similarity to UDP-D-Glc 4-epimerases. Enzyme assays of MUR4 protein expressed in the methylotropic yeast Pichia pastoris indicate that it catalyzes the 4-epimerization of UDP-D-Xyl to UDP-L-Ara, the nucleotide sugar used by glycosyltransferases in the arabinosylation of cell wall polysaccharides and wall-resident proteoglycans. Expression of MUR4-green fluorescent protein constructs in Arabidopsis revealed localization patterns consistent with targeting to the Golgi, suggesting that the MUR4 protein colocalizes with glycosyltransferases in the biosynthesis of arabinosylated cell wall components. The Arabidopsis genome encodes three putative proteins with >76% sequence identity to MUR4, which may explain why mur4 plants are not entirely deficient in the de novo synthesis of UDP-L-Ara.
Figures





References
-
- Altmann, T., Damm, B., Halfter, U., Willmitzer, L., and Morris, P.-C. (1992). Protoplast transformation and methods to create specific mutants in Arabidopsis thaliana. In Methods in Arabidopsis Research, C. Koncz, N.-H. Chua, and J. Schell, eds (Singapore: World Scientific Publishing), pp. 310–330.
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Baron-Epel, O., Gharyal, P.K., and Schindler, M. (1988). Pectins as mediators of wall porosity in soybean cells. Planta 175, 389–395. - PubMed
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris. 316, 1194–1199.
-
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, 137–144. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

