Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes
- PMID: 12566592
- PMCID: PMC141221
- DOI: 10.1105/tpc.007609
Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes
Abstract
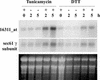
We analyzed the breadth of the unfolded protein response (UPR) in Arabidopsis using gene expression analysis with Affymetrix GeneChips. With tunicamycin and DTT as endoplasmic reticulum (ER) stress-inducing agents, we identified sets of UPR genes that were induced or repressed by both stresses. The proteins encoded by most of the upregulated genes function as part of the secretory system and comprise chaperones, vesicle transport proteins, and ER-associated degradation proteins. Most of the downregulated genes encode extracellular proteins. Therefore, the UPR may constitute a triple effort by the cell: to improve protein folding and transport, to degrade unwanted proteins, and to allow fewer secretory proteins to enter the ER. No single consensus response element was found in the promoters of the 53 UPR upregulated genes, but half of the genes contained response elements also found in mammalian UPR regulated genes. These elements are enriched from 4.5- to 15-fold in this upregulated gene set.
Figures



References
-
- Bailey, T.L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. (Menlo Park, CA: AAAI Press), pp. 28–36. - PubMed
-
- D'Amico, L., Valsasina, B., Daminati, M.G., Fabbrini, M.S., Nitti, G., Bollini, R., Ceriotti, A., and Vitale, A. (1992). Bean homologs of the mammalian glucose-regulated proteins: Induction by tunicamycin and interaction with newly synthesized seed storage proteins in the endoplasmic reticulum. Plant J. 2, 443–455. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

