Endothelial lipase is a major determinant of HDL level
- PMID: 12569160
- PMCID: PMC151857
- DOI: 10.1172/JCI16306
Endothelial lipase is a major determinant of HDL level
Abstract
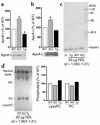
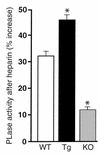
A new member of the lipase gene family, initially termed endothelial lipase (gene nomenclature, LIPG; protein, EL), is expressed in a variety of different tissues, suggesting a general role in lipid metabolism. To assess the hypothesis that EL plays a physiological role in lipoprotein metabolism in vivo, we have used gene targeting of the native murine locus and transgenic introduction of the human LIPG locus in mice to modulate the level of EL expression. Evaluation of these alleles in a C57Bl/6 background revealed an inverse relationship between HDL cholesterol level and EL expression. Fasting plasma HDL cholesterol was increased by 57% in LIPG(-/-) mice and 25% in LIPG(+/-) mice and was decreased by 19% in LIPG transgenic mice as compared with syngeneic controls. Detailed analysis of lipoprotein particle composition indicated that this increase was due primarily to an increased number of HDL particles. Phospholipase assays indicated that EL is a primary contributor to phospholipase activity in mouse. These data indicate that expression levels of this novel lipase have a significant effect on lipoprotein metabolism.
Figures



Comment in
-
Endothelial lipase: direct evidence for a role in HDL metabolism.J Clin Invest. 2003 Feb;111(3):318-21. doi: 10.1172/JCI17744. J Clin Invest. 2003. PMID: 12569156 Free PMC article. Review. No abstract available.
References
-
- Castelli WP, et al. HDL cholesterol and other lipids in coronary heart disease. The cooperative lipoprotein phenotyping study. Circulation. 1977;55:767–772. - PubMed
-
- Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am. J. Med. 1977;62:707–714. - PubMed
-
- Heller DA, de Faire U, Pedersen NL, Dahlen G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N. Engl. J. Med. 1993;328:1150–1156. - PubMed
-
- Hasstedt SJ, et al. The inheritance of high density lipoprotein cholesterol and apolipoproteins A-I and A-II. Atherosclerosis. 1984;51:21–29. - PubMed
-
- Rader DJ, Ikewaki K. Unravelling high density lipoprotein-apolipoprotein metabolism in human mutants and animal models. Curr. Opin. Lipidol. 1996;7:117–123. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

