Characterization of chitinase genes from an alkaliphilic actinomycete, Nocardiopsis prasina OPC-131
- PMID: 12571009
- PMCID: PMC143619
- DOI: 10.1128/AEM.69.2.894-900.2003
Characterization of chitinase genes from an alkaliphilic actinomycete, Nocardiopsis prasina OPC-131
Abstract
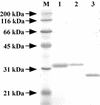
An alkaliphilic actinomycete, Nocardiopsis prasina OPC-131, secretes chitinases, ChiA, ChiB, and ChiB Delta, in the presence of chitin. The genes encoding ChiA and ChiB were cloned and sequenced. The open reading frame (ORF) of chiA encoded a protein of 336 amino acids with a calculated molecular mass of 35,257 Da. ChiA consisted of only a catalytic domain and showed a significant homology with family 18 chitinases. The chiB ORF encoded a protein of 296 amino acids with a calculated molecular mass of 31,500 Da. ChiB is a modular enzyme consisting of a chitin-binding domain type 3 (ChtBD type 3) and a catalytic domain. The catalytic domain of ChiB showed significant similarity to Streptomyces family 19 chitinases. ChiB Delta was the truncated form of ChiB lacking ChtBD type 3. Expression plasmids coding for ChiA, ChiB, and ChiB Delta were constructed to investigate the biochemical properties of these recombinant proteins. These enzymes showed pHs and temperature optima similar to those of native enzymes. ChiB showed more efficient hydrolysis of chitin and stronger antifungal activity than ChiB Delta, indicating that the ChtBD type 3 of ChiB plays an important role in the efficient hydrolysis of chitin and in antifungal activity. Furthermore, the finding of family 19 chitinase in N. prasina OPC-131 suggests that family 19 chitinases are distributed widely in actinomycetes other than the genus Streptomyces.
Figures



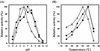
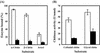

Similar articles
-
Cloning, sequencing, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain.J Bacteriol. 1997 Dec;179(23):7306-14. doi: 10.1128/jb.179.23.7306-7314.1997. J Bacteriol. 1997. PMID: 9393694 Free PMC article.
-
Molecular analysis of the gene encoding a novel cold-adapted chitinase (ChiB) from a marine bacterium, Alteromonas sp. strain O-7.J Bacteriol. 2003 Feb;185(4):1153-60. doi: 10.1128/JB.185.4.1153-1160.2003. J Bacteriol. 2003. PMID: 12562783 Free PMC article.
-
Growth of hyperthermophilic archaeon Pyrococcus furiosus on chitin involves two family 18 chitinases.Appl Environ Microbiol. 2003 Jun;69(6):3119-28. doi: 10.1128/AEM.69.6.3119-3128.2003. Appl Environ Microbiol. 2003. PMID: 12788706 Free PMC article.
-
The chitinolytic machinery of Serratia marcescens--a model system for enzymatic degradation of recalcitrant polysaccharides.FEBS J. 2013 Jul;280(13):3028-49. doi: 10.1111/febs.12181. Epub 2013 Mar 7. FEBS J. 2013. PMID: 23398882 Review.
-
Chitinolytic functions in actinobacteria: ecology, enzymes, and evolution.Appl Microbiol Biotechnol. 2018 Sep;102(17):7219-7230. doi: 10.1007/s00253-018-9149-4. Epub 2018 Jun 21. Appl Microbiol Biotechnol. 2018. PMID: 29931600 Free PMC article. Review.
Cited by
-
Marine Microbiological Enzymes: Studies with Multiple Strategies and Prospects.Mar Drugs. 2016 Sep 22;14(10):171. doi: 10.3390/md14100171. Mar Drugs. 2016. PMID: 27669268 Free PMC article. Review.
-
Isolation and identification of Streptomyces fradiae SU-1 from Thailand and protoplast transformation with the chitinase B Gene from Nocardiopsis prasina OPC-131.Curr Microbiol. 2005 Aug;51(2):116-21. doi: 10.1007/s00284-005-4402-3. Epub 2005 Jul 5. Curr Microbiol. 2005. PMID: 16010517
-
Differential chitinase activity and production within Francisella species, subspecies, and subpopulations.J Bacteriol. 2011 Jul;193(13):3265-75. doi: 10.1128/JB.00093-11. Epub 2011 Apr 29. J Bacteriol. 2011. PMID: 21531796 Free PMC article.
-
Microbial chitinases and their relevance in various industries.Folia Microbiol (Praha). 2023 Feb;68(1):29-53. doi: 10.1007/s12223-022-00999-w. Epub 2022 Aug 16. Folia Microbiol (Praha). 2023. PMID: 35972681 Review.
-
The Diversity, Metabolomics Profiling, and the Pharmacological Potential of Actinomycetes Isolated from the Estremadura Spur Pockmarks (Portugal).Mar Drugs. 2021 Dec 23;20(1):21. doi: 10.3390/md20010021. Mar Drugs. 2021. PMID: 35049876 Free PMC article.
References
-
- Al-Tai, A. M., and J.-S. Ruan. 1994. Nocardiopsis halophila sp. nov., a new halophilic actinomycete isolated from soil. Int. J. Syst. Bacteriol. 44:474-478.
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Brocq-Rousseau, D. 1904. Sur un Streptothrix. Ref. Gen. Bot. 16:20-29.
-
- Brun, E., F. Moriaud, P. Gans, M. J. Blackledge, F. Barras, and D. Marion. 1997. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry 36:16074-16086. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

