The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates
- PMID: 12571803
- PMCID: PMC1180241
- DOI: 10.1086/368264
The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates
Abstract
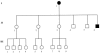
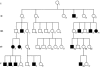
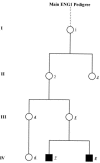
We have extended our previous analysis of the pedigree rate of control-region divergence in the human mitochondrial genome. One new germline mutation in the mitochondrial DNA (mtDNA) control region was detected among 185 transmission events (generations) from five Leber hereditary optic neuropathy (LHON) pedigrees. Pooling the LHON pedigree analyses yields a control-region divergence rate of 1.0 mutation/bp/10(6) years (Myr). When the results from eight published studies that used a similar approach were pooled with the LHON pedigree studies, totaling >2,600 transmission events, a pedigree divergence rate of 0.95 mutations/bp/Myr for the control region was obtained with a 99.5% confidence interval of 0.53-1.57. Taken together, the cumulative results support the original conclusion that the pedigree divergence rate for the control region is approximately 10-fold higher than that obtained with phylogenetic analyses. There is no evidence that any one factor explains this discrepancy, and the possible roles of mutational hotspots (rate heterogeneity), selection, and random genetic drift and the limitations of phylogenetic approaches to deal with high levels of homoplasy are discussed. In addition, we have extended our pedigree analysis of divergence in the mtDNA coding region. Finally, divergence of complete mtDNA sequences was analyzed in two tissues, white blood cells and skeletal muscle, from each of 17 individuals. In three of these individuals, there were four instances in which an mtDNA mutation was found in one tissue but not in the other. These results are discussed in terms of the occurrence of somatic mtDNA mutations.
Figures
Similar articles
-
Sequence analysis of the mitochondrial genomes from Dutch pedigrees with Leber hereditary optic neuropathy.Am J Hum Genet. 2003 Jun;72(6):1460-9. doi: 10.1086/375537. Epub 2003 May 6. Am J Hum Genet. 2003. PMID: 12736867 Free PMC article.
-
Complete mitochondrial DNA genome sequence variation of Chinese families with mutation m.3635G>A and Leber hereditary optic neuropathy.Mol Vis. 2012;18:3087-94. Epub 2012 Dec 30. Mol Vis. 2012. PMID: 23304069 Free PMC article.
-
[A study of clinical and genetic characteristics of a Leber hereditary optic neuropathy family with the heteroplasmic m.14484T>C mutation].Zhonghua Yan Ke Za Zhi. 2018 Jul 11;54(7):526-534. doi: 10.3760/cma.j.issn.0412-4081.2018.07.012. Zhonghua Yan Ke Za Zhi. 2018. PMID: 29996615 Chinese.
-
Leber Hereditary Optic Neuropathy: Exemplar of an mtDNA Disease.Handb Exp Pharmacol. 2017;240:339-376. doi: 10.1007/164_2017_2. Handb Exp Pharmacol. 2017. PMID: 28233183 Review.
-
[A review of the molecular mechanism of development of Leber hereditary optic neuropathy].Nippon Ganka Gakkai Zasshi. 2005 Apr;109(4):189-96. Nippon Ganka Gakkai Zasshi. 2005. PMID: 15859148 Review. Japanese.
Cited by
-
Time dependency of foamy virus evolutionary rate estimates.BMC Evol Biol. 2015 Jun 26;15:119. doi: 10.1186/s12862-015-0408-z. BMC Evol Biol. 2015. PMID: 26111824 Free PMC article.
-
Frequency and pattern of heteroplasmy in the control region of human mitochondrial DNA.J Mol Evol. 2008 Aug;67(2):191-200. doi: 10.1007/s00239-008-9138-9. Epub 2008 Jul 11. J Mol Evol. 2008. PMID: 18618067
-
Mutation and evolutionary rates in adélie penguins from the antarctic.PLoS Genet. 2008 Oct 3;4(10):e1000209. doi: 10.1371/journal.pgen.1000209. PLoS Genet. 2008. PMID: 18833304 Free PMC article.
-
Investigation of heteroplasmy in the human mitochondrial DNA control region: a synthesis of observations from more than 5000 global population samples.J Mol Evol. 2009 May;68(5):516-27. doi: 10.1007/s00239-009-9227-4. Epub 2009 May 1. J Mol Evol. 2009. PMID: 19407924
-
Modelling mitochondrial site polymorphisms to infer the number of segregating units and mutation rate.Biol Lett. 2009 Jun 23;5(3):397-400. doi: 10.1098/rsbl.2009.0104. Epub 2009 Mar 25. Biol Lett. 2009. PMID: 19324622 Free PMC article.
References
Electronic-Database Information
-
- Exact Binomial and Poisson Confidence Intervals, http://members.aol.com/johnp71/confint.html
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for LHON [MIM #535000]) - PubMed
References
-
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 - PubMed
-
- Bandelt H-J, Alves-Silva J, Guimaraes PEM, Santos MS, Brehm A, Pereira L, Coppa A, Larruga JM, Rengo C, Scozzari R, Torroni A, Prata MJ, Amorim A, Prado VF, Pena SDJ (2001) Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade. Ann Hum Genet 65:549–563 - PubMed
-
- Cavelier L, Jazin E, Jalonen P, Gyllensten U (2000) mtDNA substitution rate and segregation of heteroplasmy in coding and noncoding regions. Hum Genet 107:45–50 - PubMed
-
- Chinnery PF, Thorburn DR, Samuels DC, White SL, Dahl H-HM, Turnbull DM, Lightowlers RN, Howell N (2000) The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet 16:500–505 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

