The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3' processing of U2 snRNA
- PMID: 12574128
- PMCID: PMC145437
- DOI: 10.1093/emboj/cdg077
The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3' processing of U2 snRNA
Abstract
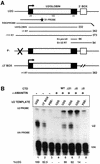
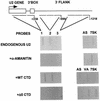
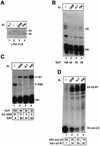
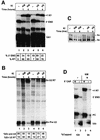
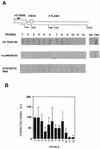
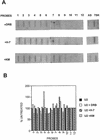
The human snRNA genes transcribed by RNA polymerase II (e.g. U1 and U2) have a characteristic TATA-less promoter containing an essential proximal sequence element. Formation of the 3' end of these non-polyadenylated RNAs requires a specialized 3' box element whose function is promoter specific. Here we show that truncation of the C-terminal domain (CTD) of RNA polymerase II and treatment of cells with CTD kinase inhibitors, including DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole), causes a dramatic reduction in proper 3' end formation of U2 transcripts. Activation of 3' box recognition by the phosphorylated CTD would be consistent with the role of phospho-CTD in mRNA processing. CTD kinase inhibitors, however, have little effect on initiation or elongation of transcription of the U2 genes, whereas elongation of transcription of the beta-actin gene is severely affected. This result highlights differences in transcription of snRNA and mRNA genes.
Figures



References
-
- Akoulitchev S., Makela,T.P., Weinberg,R.A. and Reinberg,D. (1995) Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature, 377, 557–560. - PubMed
-
- Akoulitchev S., Chuikov,S. and Reinberg,D. (2000) TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature, 407, 102–106. - PubMed
-
- Aranda A. and Proudfoot,N.J. (2001) Transcriptional termination factors for RNA polymerase II in yeast. Mol. Cell, 7, 1003–1011. - PubMed
-
- Bentley D. (2002) The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol., 14, 336–342. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

