Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY
- PMID: 12574129
- PMCID: PMC145435
- DOI: 10.1093/emboj/cdg075
Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY
Abstract
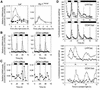
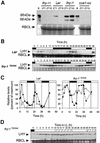
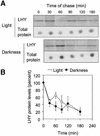
The transcription factor LHY and the related protein CCA1 perform overlapping functions in a regulatory feedback loop that is closely associated with the circadian oscillator of Arabidopsis: Overexpression of LHY abolished function of the circadian clock in constant light, but rhythmic expression of several circadian clock-regulated transcripts was observed under light- dark cycles. These oscillations correlated with high amplitude changes in LHY protein levels, caused by light-induced translation of the LHY transcript. Increases in LHY protein levels were also observed in light-grown wild-type plants, when light signals coincided with the circadian-regulated peak of LHY transcription at dawn. Unexpectedly, translational induction coincided with acute downregulation of LHY transcript levels. We suggest that the simultaneous translational induction and transcriptional repression of LHY expression play a role to narrow the peak of LHY protein synthesis at dawn and increase the robustness and accuracy of circadian oscillations. Strong phase shifting responses to light signals were observed in plants lacking function of LHY, CCA1 or both, suggesting that light-regulated expression of these proteins does not mediate entrainment of the clock to light-dark cycles.
Figures
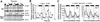



References
-
- Alabadi D., Oyama,T., Yanovsky,M.J., Harmon,F.G., Mas,P. and Kay,S.A. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science, 293, 880–883. - PubMed
-
- Alabadi D., Yanovsky,M.J., Mas,P., Harmer,S.M. and Kay,S.A. (2002) Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol., 12, 757–761. - PubMed
-
- Crosthwaite S.K., Loros,J.J. and Dunlap,J.C. (1995) Light-induced resetting of a circadian clock is mediated by a rapid increase in Frequency transcript. Cell, 81, 1003–1012. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

