An extranuclear locus of cAMP-dependent protein kinase action is necessary and sufficient for promotion of spiral ganglion neuronal survival by cAMP
- PMID: 12574406
- PMCID: PMC6741931
- DOI: 10.1523/JNEUROSCI.23-03-00777.2003
An extranuclear locus of cAMP-dependent protein kinase action is necessary and sufficient for promotion of spiral ganglion neuronal survival by cAMP
Abstract
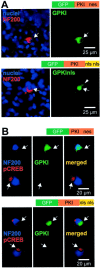
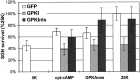
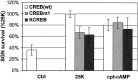
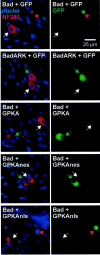
We showed previously that cAMP is a survival-promoting stimulus for cultured postnatal rat spiral ganglion neurons (SGNs) and that depolarization promotes SGN survival in part via recruitment of cAMP signaling. We here investigate the subcellular locus of cAMP prosurvival signaling. Transfection of GPKI, a green fluorescent protein (GFP)-tagged cAMP-dependent protein kinase (PKA) inhibitor, inhibits the ability of the permeant cAMP analog cpt-cAMP [8-(4-chlorophenylthio)-cAMP] to promote survival, indicating that PKA activity is necessary. Transfection of GFP-tagged PKA (GPKA) is sufficient to promote SGN survival, but restriction of GPKA to the nucleus by addition of a nuclear localization signal (GPKAnls) almost completely abrogates its prosurvival effect. In contrast, GPKA targeted to the extranuclear cytoplasm by addition of a nuclear export signal (GPKAnes) promotes SGN survival as effectively as does GPKA. Moreover, GPKI targeted to the nucleus lacks inhibitory effect on SGN survival attributable to cpt-cAMP or depolarization. These data indicate an extranuclear target of PKA for promotion of neuronal survival. Consistent with this, we find that dominant-inhibitory CREB mutants inhibit the prosurvival effect of depolarization but not that of cpt-cAMP. SGN survival is compromised by overexpression of the proapoptotic regulator Bad, previously shown to be phosphorylated in the cytoplasm by PKA. This Bad-induced apoptosis is prevented by cpt-cAMP or by cotransfection of GPKA or of GPKAnes but not of GPKAnls. Thus, cAMP prevents SGN death through a cytoplasmic as opposed to nuclear action, and inactivation of Bad proapoptotic function is a mechanism by which PKA can prevent neuronal death.
Figures





References
-
- Antoni FA. Molecular diversity of cyclic AMP signalling. Front Neuroendocrinol. 2000;21:103–132. - PubMed
-
- Bacskai BJ, Hochner B, Mahaut-Smith M, Adams SR, Kaang BK, Kandel ER, Tsien RY. Spatially resolved dynamics of cAMP and protein kinase A subunits in Aplysia sensory neurons. Science. 1993;260:222–226. - PubMed
-
- Billiard J, Grewal SS, Lukaesko L, Stork PJS, Rotwein P. Hormonal control of insulin-like growth factor-I gene transcription in human osteoblasts: dual actions of cAMP-dependent protein kinase on CCAAT/enhancer binding protein δ. J Biol Chem. 2001;276:31238–31246. - PubMed
-
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
