Repression of the HIV-1 5' LTR promoter and inhibition of HIV-1 replication by using engineered zinc-finger transcription factors
- PMID: 12574502
- PMCID: PMC149881
- DOI: 10.1073/pnas.252770699
Repression of the HIV-1 5' LTR promoter and inhibition of HIV-1 replication by using engineered zinc-finger transcription factors
Abstract
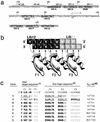
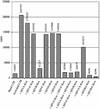
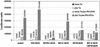
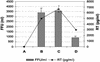
Zinc finger domains are small DNA-binding modules that can be engineered to bind desired target sequences. Functional transcription factors can be made from these DNA-binding modules, by fusion with an appropriate effector domain. In this study, eight three-zinc-finger proteins (ZFPs) that bound HIV-1 sequences in vitro were engineered into transcription repressors by linking them to the Krüppel-associated box (KRAB) repressor domain (KOX1). One protein, ZFP HIVB-KOX, which bound to a 9-bp region overlapping two Sp1 sites, was found to repress a Tat-activated 5' LTR cellular HIV-reporter assay to almost basal levels. A related six-finger protein, HIVBA'-KOX, was made to target all three Sp1 sites in the 5' LTR promoter and efficiently inhibited both basal and Tat-activated transcription in unstimulated and mitogen-stimulated T cells. In contrast, a combination of two unlinked three-finger ZFPs, HIVA'-KOX and HIVB-KOX, which bind over the same region of DNA, resulted in less effective repression. Finally, HIVBA'-KOX was tested for its capacity to block viral replication in a cellular infection assay using the HIV-1 HXB2 strain. This ZFP was found to inhibit HIV-1 replication by 75% compared with control constructs, thus demonstrating the potential of this approach for antiviral therapy.
Figures

Similar articles
-
Enhancement of the basal-level activity of HIV-1 long terminal repeat by HIV-1 nucleocapsid protein.Virology. 2000 Mar 15;268(2):251-63. doi: 10.1006/viro.2000.0194. Virology. 2000. PMID: 10704334
-
BCL6 can repress transcription from the human immunodeficiency virus type I promoter/enhancer region.Genes Chromosomes Cancer. 1997 May;19(1):14-21. Genes Chromosomes Cancer. 1997. PMID: 9135990
-
[Construction of a SV40 promoter specific artificial transcription factor].Sheng Wu Gong Cheng Xue Bao. 2003 Sep;19(5):608-12. Sheng Wu Gong Cheng Xue Bao. 2003. PMID: 15969093 Chinese.
-
Step out of the groove: epigenetic gene control systems and engineered transcription factors.Adv Genet. 2006;56:163-204. doi: 10.1016/S0065-2660(06)56005-5. Adv Genet. 2006. PMID: 16735158 Review.
-
Drug discovery with engineered zinc-finger proteins.Nat Rev Drug Discov. 2003 May;2(5):361-8. doi: 10.1038/nrd1087. Nat Rev Drug Discov. 2003. PMID: 12750739 Review.
Cited by
-
A minimal region of the HSP90AB1 promoter is suitable for ubiquitous expression in different somatic tissues with applicability for gene therapy.Front Mol Biosci. 2023 Apr 17;10:1175407. doi: 10.3389/fmolb.2023.1175407. eCollection 2023. Front Mol Biosci. 2023. PMID: 37138658 Free PMC article.
-
Broadly active zinc finger protein-guided transcriptional activation of HIV-1.Mol Ther Methods Clin Dev. 2020 Oct 27;20:18-29. doi: 10.1016/j.omtm.2020.10.018. eCollection 2021 Mar 12. Mol Ther Methods Clin Dev. 2020. PMID: 33335944 Free PMC article.
-
The zinc finger DNA-binding domain of K-RBP plays an important role in regulating Kaposi's sarcoma-associated herpesvirus RTA-mediated gene expression.Virology. 2009 Sep 1;391(2):221-31. doi: 10.1016/j.virol.2009.06.014. Epub 2009 Jul 9. Virology. 2009. PMID: 19592062 Free PMC article.
-
Artificial 64-Residue HIV-1 Enhancer-Binding Peptide Is a Potent Inhibitor of Viral Replication in HIV-1-Infected Cells.Adv Virol. 2011;2011:165871. doi: 10.1155/2011/165871. Epub 2011 Sep 19. Adv Virol. 2011. PMID: 22312334 Free PMC article.
-
Designed zinc finger protein interacting with the HIV-1 integrase recognition sequence at 2-LTR-circle junctions.Protein Sci. 2009 Nov;18(11):2219-30. doi: 10.1002/pro.233. Protein Sci. 2009. PMID: 19701937 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

