Selective incorporation of influenza virus RNA segments into virions
- PMID: 12574509
- PMCID: PMC149948
- DOI: 10.1073/pnas.0437772100
Selective incorporation of influenza virus RNA segments into virions
Abstract
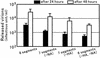
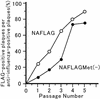
The genome of influenza A virus is comprised of eight viral RNA (vRNA) segments. Although the products of all eight vRNA segments must be present for viral replication, little is known about the mechanism(s) responsible for incorporation of these segments into virions. Two models have been proposed for the generation of infectious virions containing eight vRNA segments. The random-incorporation model assumes a common structural feature in all the vRNAs, enabling any combination of vRNAs to be incorporated randomly into virions. The selective-incorporation model predicts the presence of specific structures in each vRNA segment, leading to the incorporation of a set of eight vRNA segments into virions. Here we demonstrate that eight different vRNA segments must be present for efficient virion formation and that sequences within the coding region of (and thus unique to) the neuraminidase vRNA possess a signal that drives incorporation of this segment into virions. These findings indicate a unique contribution from individual vRNA segments and thus suggest a selective (rather than random) mechanism of vRNA recruitment into virions. The neuraminidase vRNA incorporation signal and others yet to be identified should provide attractive targets for the attenuation of influenza viruses in vaccine production and the design of new antiviral drugs.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

