DNA duplex-quadruplex exchange as the basis for a nanomolecular machine
- PMID: 12574521
- PMCID: PMC149873
- DOI: 10.1073/pnas.0335459100
DNA duplex-quadruplex exchange as the basis for a nanomolecular machine
Abstract
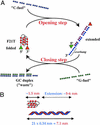
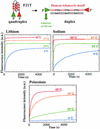
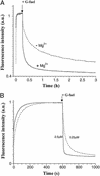
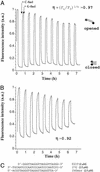
There is currently great interest in the design of nanodevices that are capable of performing linear or rotary movements. Protein molecular machines are abundant in biology but it has recently been proposed that nucleic acids could also act as nanomolecular machines in model systems. Several types of movements have been described with DNA machines: rotation and "scissors-like" opening and closing. Here we show a nanomachine that is capable of an extension-contraction movement. The simple and robust device described here is composed of a single 21-base oligonucleotide and relies on a duplex-quadruplex equilibrium that may be fueled by the sequential addition of DNA single strands, generating a DNA duplex as a by-product. The interconversion between two well defined topological states induces a 5-nm two-stroke, linear motor-type movement, which is detected by fluorescence resonance energy transfer spectroscopy.
Figures

References
-
- Balzani V V, Credi A, Raymo F M, Stoddart J F. Angew Chem Int Ed Engl. 2000;39:3348–3391. - PubMed
-
- Alberts B. Cell. 1998;92:291–294. - PubMed
-
- Mao C D, Sun W Q, Shen Z Y, Seeman N C. Nature. 1999;397:144–146. - PubMed
-
- Yurke B, Tuberfield A J, Mills A P, Jr, Simmel F C, Neunmann J L. Nature. 2000;406:605–608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

